A Re-Examination of the Molecular Systematics and Phylogeography of Taxa of the Peromyscus Aztecus Species Group, with Comments on the Distribution of P
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Special Publications Museum of Texas Tech University Number 63 18 September 2014
Special Publications Museum of Texas Tech University Number 63 18 September 2014 List of Recent Land Mammals of Mexico, 2014 José Ramírez-Pulido, Noé González-Ruiz, Alfred L. Gardner, and Joaquín Arroyo-Cabrales.0 Front cover: Image of the cover of Nova Plantarvm, Animalivm et Mineralivm Mexicanorvm Historia, by Francisci Hernández et al. (1651), which included the first list of the mammals found in Mexico. Cover image courtesy of the John Carter Brown Library at Brown University. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 63 List of Recent Land Mammals of Mexico, 2014 JOSÉ RAMÍREZ-PULIDO, NOÉ GONZÁLEZ-RUIZ, ALFRED L. GARDNER, AND JOAQUÍN ARROYO-CABRALES Layout and Design: Lisa Bradley Cover Design: Image courtesy of the John Carter Brown Library at Brown University Production Editor: Lisa Bradley Copyright 2014, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (nsrl.ttu.edu). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 18 September 2014 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 63 Series Editor: Robert J. -

Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca
Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca John F. Lamoreux, Meghan W. McKnight, and Rodolfo Cabrera Hernandez Occasional Paper of the IUCN Species Survival Commission No. 53 Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca John F. Lamoreux, Meghan W. McKnight, and Rodolfo Cabrera Hernandez Occasional Paper of the IUCN Species Survival Commission No. 53 The designation of geographical entities in this book, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The views expressed in this publication do not necessarily reflect those of IUCN or other participating organizations. Published by: IUCN, Gland, Switzerland Copyright: © 2015 International Union for Conservation of Nature and Natural Resources Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. Reproduction of this publication for resale or other commercial purposes is prohibited without prior written permission of the copyright holder. Citation: Lamoreux, J. F., McKnight, M. W., and R. Cabrera Hernandez (2015). Amphibian Alliance for Zero Extinction Sites in Chiapas and Oaxaca. Gland, Switzerland: IUCN. xxiv + 320pp. ISBN: 978-2-8317-1717-3 DOI: 10.2305/IUCN.CH.2015.SSC-OP.53.en Cover photographs: Totontepec landscape; new Plectrohyla species, Ixalotriton niger, Concepción Pápalo, Thorius minutissimus, Craugastor pozo (panels, left to right) Back cover photograph: Collecting in Chamula, Chiapas Photo credits: The cover photographs were taken by the authors under grant agreements with the two main project funders: NGS and CEPF. -

Mammal Species Native to the USA and Canada for Which the MIL Has an Image (296) 31 July 2021
Mammal species native to the USA and Canada for which the MIL has an image (296) 31 July 2021 ARTIODACTYLA (includes CETACEA) (38) ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BALAENIDAE - bowheads and right whales 1. Balaena mysticetus – Bowhead Whale BALAENOPTERIDAE -rorqual whales 1. Balaenoptera acutorostrata – Common Minke Whale 2. Balaenoptera borealis - Sei Whale 3. Balaenoptera brydei - Bryde’s Whale 4. Balaenoptera musculus - Blue Whale 5. Balaenoptera physalus - Fin Whale 6. Eschrichtius robustus - Gray Whale 7. Megaptera novaeangliae - Humpback Whale BOVIDAE - cattle, sheep, goats, and antelopes 1. Bos bison - American Bison 2. Oreamnos americanus - Mountain Goat 3. Ovibos moschatus - Muskox 4. Ovis canadensis - Bighorn Sheep 5. Ovis dalli - Thinhorn Sheep CERVIDAE - deer 1. Alces alces - Moose 2. Cervus canadensis - Wapiti (Elk) 3. Odocoileus hemionus - Mule Deer 4. Odocoileus virginianus - White-tailed Deer 5. Rangifer tarandus -Caribou DELPHINIDAE - ocean dolphins 1. Delphinus delphis - Common Dolphin 2. Globicephala macrorhynchus - Short-finned Pilot Whale 3. Grampus griseus - Risso's Dolphin 4. Lagenorhynchus albirostris - White-beaked Dolphin 5. Lissodelphis borealis - Northern Right-whale Dolphin 6. Orcinus orca - Killer Whale 7. Peponocephala electra - Melon-headed Whale 8. Pseudorca crassidens - False Killer Whale 9. Sagmatias obliquidens - Pacific White-sided Dolphin 10. Stenella coeruleoalba - Striped Dolphin 11. Stenella frontalis – Atlantic Spotted Dolphin 12. Steno bredanensis - Rough-toothed Dolphin 13. Tursiops truncatus - Common Bottlenose Dolphin MONODONTIDAE - narwhals, belugas 1. Delphinapterus leucas - Beluga 2. Monodon monoceros - Narwhal PHOCOENIDAE - porpoises 1. Phocoena phocoena - Harbor Porpoise 2. Phocoenoides dalli - Dall’s Porpoise PHYSETERIDAE - sperm whales Physeter macrocephalus – Sperm Whale TAYASSUIDAE - peccaries Dicotyles tajacu - Collared Peccary CARNIVORA (48) CANIDAE - dogs 1. Canis latrans - Coyote 2. -

Wildlife Ruby Lake Natillntllwildlife Refuge
I 49. 44/2: R 82/3/993 P RLE Wildlife Ruby lake NatillntllWildlife Refuge ZIMMERMAN LIBRARY UNIV. OF NEW MEXteo FEB 1 0 1994 U.S. Regional Depos1to A Refuge for Nesting and Migrating Waterfowl and Other Wildlife The Habitat Ruby Lake National Wildlife Refuge was established in The refuge, at an elevation of 6,000 feet, consists of an 1938. It encompasses 37,632 acres at the south end of extensive bulrush marsh interspersed with pockets of Ruby Valley. This land was once covered by a 200 foot open water. Fish are abundant. Islands scattered deep, 300,800-acre lake known as Franklin Lake. Today throughout provide good nesting habitat for many bird 12,000 acres of marsh remain on the refuge. Just north of species. the refuge, a 15,000-acre seasonal wetland is now referred to as Franklin Lake. Over 200 springs flow into the marsh along its west border _...)/ creating riparian habitat which is used by many songbirds, To Elko �� and Welle snipe, rail and small mammals. They also provide a water FRANKLIN source for larger mammals. With slight increases in LAKE elevation, wet meadows gradate into grasslands and sagebrush-rabbitbrush habitat. Pinon pines and juniper cover the slopes of the Ruby Mountains that rise to 11,000 feet along the west side of the refuge. Canyons provide habitat for a variety of wildlife. Rock cliffs provide raptors with nesting and perching sites. A mountainside of dead trees, home for ROAD cavity dwelling birds, was the result of a 1979 wildfire. BRESSMAN CABIN LOOP MAIN BOAT LANDING -4,__,,� ·�I! I N � 0 3 Miles 0 2 4 Kilometer� RANCH dead pinon tree General Key BIRDS bam ,wallow � Season 6 The following bird list includes 207 species observed on Sp - Spring (March through May) or near the refuge. -

TESIS: Ámbito Hogareño Y Selección De Hábitat De Reithrodontomys
UNIVERSIDAD NACIONAL AUTÓNOMA DE MÉXICO FACULTAD DE CIENCIAS Ámbito hogareño y selección de hábitat de Reithrodontomys microdon (Cricetidae: Neotominae) T E S I S QUE PARA OBTENER EL TÍTULO DE: B I Ó L O G A P R E S E N T A : Tania Marines Macías DIRECTORA DE TESIS: Dra. Livia Socorro León Paniagua 2014 UNAM – Dirección General de Bibliotecas Tesis Digitales Restricciones de uso DERECHOS RESERVADOS © PROHIBIDA SU REPRODUCCIÓN TOTAL O PARCIAL Todo el material contenido en esta tesis esta protegido por la Ley Federal del Derecho de Autor (LFDA) de los Estados Unidos Mexicanos (México). El uso de imágenes, fragmentos de videos, y demás material que sea objeto de protección de los derechos de autor, será exclusivamente para fines educativos e informativos y deberá citar la fuente donde la obtuvo mencionando el autor o autores. Cualquier uso distinto como el lucro, reproducción, edición o modificación, será perseguido y sancionado por el respectivo titular de los Derechos de Autor. 1. Datos del alumno Marines Macías Tania 26155080 Universidad Nacional Autónoma de México Facultad de Ciencias Biología 305292504 2. Datos del tutor Dra. Livia Socorro León Paniagua 3. Datos del sinodal 1 Dr. Cano Santana Zenón 4. Datos del sinodal 2 Dr. José Jaime Zúñiga Vega 5. Datos del sinodal 3 Dr. Ávila Flores Rafael 6. Datos del sinodal 4 M. en B. Zamira Anahí Ávila Valle 7. Datos del trabajo escrito Ámbito hogareño y selección de hábitat de Reithrodontomys microdon (Cricetidae: Neotominae) 46 p 2014 Agradecimientos La presente tesis fue desarrollada durante el curso del Taller “Faunística, sistemática y biogeografía de vertebrados terrestres de México”, en el Departamento de Biología Evolutiva de la Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM). -
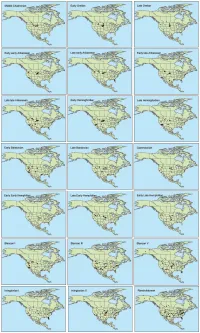
Supporting Files
Table S1. Summary of Special Emissions Report Scenarios (SERs) to which we fit climate models for extant mammalian species. Mean Annual Temperature Standard Scenario year (˚C) Deviation Standard Error Present 4.447 15.850 0.057 B1_low 2050s 5.941 15.540 0.056 B1 2050s 6.926 15.420 0.056 A1b 2050s 7.602 15.336 0.056 A2 2050s 8.674 15.163 0.055 A1b 2080s 7.390 15.444 0.056 A2 2080s 9.196 15.198 0.055 A2_top 2080s 11.225 14.721 0.053 Table S2. List of mammalian taxa included and excluded from the species distribution models. -

The Neotropical Region Sensu the Areas of Endemism of Terrestrial Mammals
Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Supplementary material The Neotropical region sensu the areas of endemism of terrestrial mammals Elkin Alexi Noguera-UrbanoA,B,C,D and Tania EscalanteB APosgrado en Ciencias Biológicas, Unidad de Posgrado, Edificio A primer piso, Circuito de Posgrados, Ciudad Universitaria, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. BGrupo de Investigación en Biogeografía de la Conservación, Departamento de Biología Evolutiva, Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. CGrupo de Investigación de Ecología Evolutiva, Departamento de Biología, Universidad de Nariño, Ciudadela Universitaria Torobajo, 1175-1176 Nariño, Colombia. DCorresponding author. Email: [email protected] Page 1 of 18 Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Table S1. List of taxa processed Number Taxon Number Taxon 1 Abrawayaomys ruschii 55 Akodon montensis 2 Abrocoma 56 Akodon mystax 3 Abrocoma bennettii 57 Akodon neocenus 4 Abrocoma boliviensis 58 Akodon oenos 5 Abrocoma budini 59 Akodon orophilus 6 Abrocoma cinerea 60 Akodon paranaensis 7 Abrocoma famatina 61 Akodon pervalens 8 Abrocoma shistacea 62 Akodon philipmyersi 9 Abrocoma uspallata 63 Akodon reigi 10 Abrocoma vaccarum 64 Akodon sanctipaulensis 11 Abrocomidae 65 Akodon serrensis 12 Abrothrix 66 Akodon siberiae 13 Abrothrix andinus 67 Akodon simulator 14 Abrothrix hershkovitzi 68 Akodon spegazzinii 15 Abrothrix illuteus -

Redalyc.SMALL MAMMAL COMMUNITIES in the SIERRA DE
Mastozoología Neotropical ISSN: 0327-9383 [email protected] Sociedad Argentina para el Estudio de los Mamíferos Argentina Matson, John O.; Ordóñez-Garza, Nicté; Bulmer, Walter; Eckerlin, Ralph P. SMALL MAMMAL COMMUNITIES IN THE SIERRA DE LOS CUCHUMATANES, HUEHUETENANGO, GUATEMALA Mastozoología Neotropical, vol. 19, núm. 1, enero-junio, 2012, pp. 71-84 Sociedad Argentina para el Estudio de los Mamíferos Tucumán, Argentina Available in: http://www.redalyc.org/articulo.oa?id=45723408007 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Mastozoología Neotropical, 19(1):71-84, Mendoza, 2012 ISSN 0327-9383 ©SAREM, 2012 Versión on-line ISSN 1666-0536 http://www.sarem.org.ar SMALL MAMMAL COMMUNITIES IN THE SIERRA DE LOS CUCHUMATANES, HUEHUETENANGO, GUATEMALA John O. Matson1, Nicté Ordóñez-Garza2, Walter Bulmer3, and Ralph P. Eckerlin3 1 Department of Biological Sciences, San Jose State University, San Jose, CA 95192-0100 [Corres- pondence: <[email protected]>]. 2 Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409-3131. 3 Division of Natural Sciences and Mathematics, Northern Virginia Com- munity College, Annandale, VA 22003-3796. ABSTRACT: Very little is known concerning small mammal ecology and their distribution in the highlands of Guatemala. Small mammals were trapped from five different cloud forests in the Sierra de los Cuchumatanes, Huehuetenango, Guatemala. Cloud forest elevations ranged from 2600 m to 3350 m. Most sites had evidence of human disturbance with only Cerro Bobí having a relatively pristine forest. -

Chapter 5 a New Species of Reithrodontomys, Subgenus
Chapter 5 A New Species of Reithrodontomys, Subgenus Aporodon (Cricetidae: Neotominae), from the Highlands of Costa Rica, with Comments on Costa Rican and Panamanian Reithrodontomys ALFRED L. GARDNER1 AND MICHAEL D. CARLETON2 ABSTRACT A new species of the rodent genus Reithrodontomys (Cricetidae: Neotominae) is described from Cerro Asuncio´n in the western Cordillera de Talamanca, Costa Rica. The long tail, elongate rostrum, bulbous braincase, and complex molars of the new species associate it with members of the subgenus Aporodon, tenuirostris species group. In its diminutive size and aspects of cranial shape, the new species (Reithrodontomys musseri, sp. nov.) most closely resembles R. microdon, a form known from highlands in Guatemala and Chiapas, Mexico. In the courseof differentially diagnosing the new species, we necessarily reviewed the Costa Rican and Panamanian subspecies of R. mexicanus based on morphological comparisons, study of paratypes and vouchers used in recent molecular studies, and morphometric analyses. We recognize Reithrodontomys cherrii (Allen, 1891) and R. garichensis Enders and Pearson, 1940, as valid species, and allocate R. mexicanus potrerograndei Goodwin, 1945, as a subjective synonym of R. brevirostris Goodwin, 1943. Critical review of museum specimens collected subsequent to Hooper’s (1952) revision is needed and would do much to improve understanding of Reithrodontomys taxonomy and distribution in Middle America. INTRODUCTION unknown to the senior author at the time. Among the mammals collected in Costa Several days of trapping at the collecting site Rica during 1966 and 1967, when Gardner and elsewhere on the Cerro Buenavista massif held an appointment with the Louisiana State over the next five months failed to produce University International Center for Medical additional specimens. -

(Mammalia) from Durango, Western Mexico
Check List 9(2): 313–322, 2013 © 2013 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution A checklist of the mammals (Mammalia) from Durango, PECIES S western Mexico OF Diego F. García-Mendoza * and Celia López-González ISTS L CIIDIR Unidad Durango, Instituto Politécnico Nacional, Calle Sigma 119, Fraccionamiento 20 de Noviembre II, Durango, Durango, México 34220. * Corresponding author. E-mail: [email protected] Abstract: An updated list of the mammals of Durango state, Mexico was built from literature records and Museum specimens. A total of 139 species have been recorded, representing 28.3 % of the Mexican terrestrial mammals, and 25.1 % species more compared to the previous account. Two species have been extirpated from the state, 23 are endemic to Mexico. Four major ecoregions have been previously defined for the state, Arid, Valleys, Sierra, and Quebradas. Species richness is ofhighest the largest at the diversitiesQuebradas, of a mammalstropical ecorregion, of the country, whereas conservation the aridlands efforts are arethe minimal,least species-rich. and the current The Sierra protected has the areas highest do number of endemic species (11) followed by Quebradas (7), Valleys and Arid (3). Despite the fact that Durango harbors one in practice already existing plans to protect Durango’s unique biodiversity. not include the most species-rich regions. The current rate of anthropogenic modification in the state makes urgent to put Introduction the only treatise on the mammalian fauna of the state. Mexico harbors one of the most diverse mammalian Following this work, a number of papers on the taxonomy fauna (527 species, 488 terrestrial), surpassed only by and distribution of Durango mammals have been published Brazil and Indonesia. -

Approximately 220 Species of Wild Mammals Occur in California And
Mammals pproximately 220 species of wild mammals occur in California and the surrounding waters (including introduced species, but not domestic species Asuch as house cats). Amazingly, the state of California has about half of the total number of species that occur on the North American continent (about 440). In part, this diversity reflects the sheer number of different habitats available throughout the state, including alpine, desert, coniferous forest, grassland, oak woodland, and chaparral habitat types, among others (Bakker 1984, Schoenherr 1992, Alden et al. 1998). About 17 mammal species are endemic to California; most of these are kangaroo rats, chipmunks, and squirrels. Nearly 25% of California’s mammal species are either known or suspected to occur at Quail Ridge (Appendix 9). Species found at Quail Ridge are typical of both the Northwestern California and Great Central Valley mammalian faunas. Two California endemics, the Sonoma chipmunk (see Species Accounts for scientific names) and the San Joaquin pocket mouse, are known to occur at Quail Ridge. None of the mammals at Quail Ridge are listed as threatened or endangered by either the state or federal governments, although Townsend’s big-eared bat, which is suspected to occur at Quail Ridge, is a state-listed species of special concern. Many mammal species are nocturnal, fossorial, fly, or are otherwise difficult to observe. However, it is still possible to detect the presence of mammals at Quail Ridge, both visually and by observation of their tracks, scat, and other sign. The mammals most often seen during the day are mule deer and western gray squirrels. -

Over 40% of All Mammal Species in the Next 2 Labs
Rodents Class Rodentia 5 (depends) Suborders 33 (maybe more) Families about 481 genera, 2277+ species Over 40% of all mammal species in the next 2 labs Sciuromorpha: squirrels, dormice, mountain beaver, and relatives Castorimorpha: beavers, gophers, kangaroo rats, pocket mice, and relatives Myomorpha: mice, rats, gerbils, jerboas, and relatives Anomaluromorpha: scaly-tailed squirrels and springhares Hystricomorpha: hystricognath rodents...lots of South American and African species, mostly Because rodents are such a Why rodents are evil... diverse and speciose group, their higher-level taxonomy keeps being revised. Hard to keep up! In recent decades, there have been 2, 3, 4 or 5 Suborders, depending on the revision, and Families keep getting pooled and split. We’ll just focus on some of the important Families and leave their relationships to future generations. They are a diverse and Why rodents are fun... speciose group, occur in just about every kind of habitat and climate, and show the broadest ecological diversity of any group of mammals. There are terrestrial, arboreal, scansorial, subterranean, and semiaquatic rodents. There are solitary, pair-forming, and social rodents. There are plantigrade, cursorial, You could spend your whole fossorial, bipedal, swimming life studying this group! and gliding rodents. (Some do.) General characteristics of rodents •Specialized ever-growing, self-sharpening incisors (2 upper, 2 lower) separated from cheek teeth by diastema; no canines •Cheek teeth may be ever-growing or rooted, but show a variety of cusp patterns, often with complex loops and folds of enamel and dentine reflecting the diet; cusp patterns also often useful taxonomically •Mostly small, average range of body size is 20-100 g, but some can get pretty large (capybara is largest extant species, may reach 50 kg) •Mostly herbivorous (including some specialized as folivores and granivores) or omnivorous •Females with duplex uterus, baculum present in males •Worldwide distribution, wide range of habitats and ecologies And now, on to a few Families..