Spatial Patterns of Phylogenetic Diversity of Mexican Mammals for Biodiversity Conservation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Special Publications Museum of Texas Tech University Number 63 18 September 2014
Special Publications Museum of Texas Tech University Number 63 18 September 2014 List of Recent Land Mammals of Mexico, 2014 José Ramírez-Pulido, Noé González-Ruiz, Alfred L. Gardner, and Joaquín Arroyo-Cabrales.0 Front cover: Image of the cover of Nova Plantarvm, Animalivm et Mineralivm Mexicanorvm Historia, by Francisci Hernández et al. (1651), which included the first list of the mammals found in Mexico. Cover image courtesy of the John Carter Brown Library at Brown University. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 63 List of Recent Land Mammals of Mexico, 2014 JOSÉ RAMÍREZ-PULIDO, NOÉ GONZÁLEZ-RUIZ, ALFRED L. GARDNER, AND JOAQUÍN ARROYO-CABRALES Layout and Design: Lisa Bradley Cover Design: Image courtesy of the John Carter Brown Library at Brown University Production Editor: Lisa Bradley Copyright 2014, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (nsrl.ttu.edu). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 18 September 2014 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 63 Series Editor: Robert J. -
Supporting Files
Table S1. Summary of Special Emissions Report Scenarios (SERs) to which we fit climate models for extant mammalian species. Mean Annual Temperature Standard Scenario year (˚C) Deviation Standard Error Present 4.447 15.850 0.057 B1_low 2050s 5.941 15.540 0.056 B1 2050s 6.926 15.420 0.056 A1b 2050s 7.602 15.336 0.056 A2 2050s 8.674 15.163 0.055 A1b 2080s 7.390 15.444 0.056 A2 2080s 9.196 15.198 0.055 A2_top 2080s 11.225 14.721 0.053 Table S2. List of mammalian taxa included and excluded from the species distribution models. -

The Neotropical Region Sensu the Areas of Endemism of Terrestrial Mammals
Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Supplementary material The Neotropical region sensu the areas of endemism of terrestrial mammals Elkin Alexi Noguera-UrbanoA,B,C,D and Tania EscalanteB APosgrado en Ciencias Biológicas, Unidad de Posgrado, Edificio A primer piso, Circuito de Posgrados, Ciudad Universitaria, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. BGrupo de Investigación en Biogeografía de la Conservación, Departamento de Biología Evolutiva, Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM), 04510 Mexico City, Mexico. CGrupo de Investigación de Ecología Evolutiva, Departamento de Biología, Universidad de Nariño, Ciudadela Universitaria Torobajo, 1175-1176 Nariño, Colombia. DCorresponding author. Email: [email protected] Page 1 of 18 Australian Systematic Botany, 2017, 30, 470–484 ©CSIRO 2017 doi:10.1071/SB16053_AC Table S1. List of taxa processed Number Taxon Number Taxon 1 Abrawayaomys ruschii 55 Akodon montensis 2 Abrocoma 56 Akodon mystax 3 Abrocoma bennettii 57 Akodon neocenus 4 Abrocoma boliviensis 58 Akodon oenos 5 Abrocoma budini 59 Akodon orophilus 6 Abrocoma cinerea 60 Akodon paranaensis 7 Abrocoma famatina 61 Akodon pervalens 8 Abrocoma shistacea 62 Akodon philipmyersi 9 Abrocoma uspallata 63 Akodon reigi 10 Abrocoma vaccarum 64 Akodon sanctipaulensis 11 Abrocomidae 65 Akodon serrensis 12 Abrothrix 66 Akodon siberiae 13 Abrothrix andinus 67 Akodon simulator 14 Abrothrix hershkovitzi 68 Akodon spegazzinii 15 Abrothrix illuteus -

(Mammalia) from Durango, Western Mexico
Check List 9(2): 313–322, 2013 © 2013 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution A checklist of the mammals (Mammalia) from Durango, PECIES S western Mexico OF Diego F. García-Mendoza * and Celia López-González ISTS L CIIDIR Unidad Durango, Instituto Politécnico Nacional, Calle Sigma 119, Fraccionamiento 20 de Noviembre II, Durango, Durango, México 34220. * Corresponding author. E-mail: [email protected] Abstract: An updated list of the mammals of Durango state, Mexico was built from literature records and Museum specimens. A total of 139 species have been recorded, representing 28.3 % of the Mexican terrestrial mammals, and 25.1 % species more compared to the previous account. Two species have been extirpated from the state, 23 are endemic to Mexico. Four major ecoregions have been previously defined for the state, Arid, Valleys, Sierra, and Quebradas. Species richness is ofhighest the largest at the diversitiesQuebradas, of a mammalstropical ecorregion, of the country, whereas conservation the aridlands efforts are arethe minimal,least species-rich. and the current The Sierra protected has the areas highest do number of endemic species (11) followed by Quebradas (7), Valleys and Arid (3). Despite the fact that Durango harbors one in practice already existing plans to protect Durango’s unique biodiversity. not include the most species-rich regions. The current rate of anthropogenic modification in the state makes urgent to put Introduction the only treatise on the mammalian fauna of the state. Mexico harbors one of the most diverse mammalian Following this work, a number of papers on the taxonomy fauna (527 species, 488 terrestrial), surpassed only by and distribution of Durango mammals have been published Brazil and Indonesia. -

THOMAS E. LACHER, JR., Ph.D
CURRICULUM VITAE THOMAS E. LACHER, JR., Ph.D. Professor Department of Ecology and Conservation Biology TAMU 2258 Texas A&M University College Station, TX 77843-2258 Main Phone: (979) 845-5777; Fax: (979) 845-3786 E-mail: [email protected] EDUCATION B.S.: April, 1972, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA, minors in Chemistry and English, overall GPA (137 semester credits)-3.45. Undergraduate honors: Pennsylvania State Scholarship, Senatorial Scholarship, Honors Major in Biology, Cum Laude graduate, Dean's List - 8 semesters. Ph.D.: April, 1980, Department of Biological Sciences, Section of Ecology, Evolution and Systematics, University of Pittsburgh, Pittsburgh, PA, overall GPA (61 semester credits)-3.93. Dissertation: The Comparative Social Behavior of Kerodon rupestris and Galea spixii in the Xeric Caatinga of Northeastern Brazil - Michael A. Mares, major advisor. Language proficiency: Read Italian (basic); Read and speak French (basic); Read, write, understand, and speak Spanish (proficient); Read, write, understand, and speak English and Portuguese (fluent) EMPLOYMENT HISTORY AND TEACHING EXPERIENCE Research/Teaching/Tenure Track Positions 2007-present Full Professor, Department of Ecology and Conservation Biology (formerly Wildlife and Fisheries Sciences), Texas A&M University. Core Faculty in the Interdisciplinary Doctoral Program in Ecology and Evolutionary Biology (EEB) and Core Faculty of the NSF IGERT in Applied Biodiversity Science. Teaching experience includes Dominica Tropical and Field Biology, Wildlife and Fisheries Conservation, and Mammalogy (all UG) and the Ecosystem Ecology Module in the EEB Core and Applied Biodiversity Science I (Grad). Also Director (2019 – present), Center for Coffee Research and Education, Norman Borlaug Institute for International Agriculture, Texas A&M Agrilife Research. -

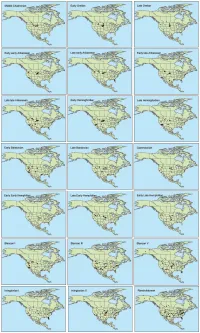
A Global-Scale Evaluation of Mammalian Exposure and Vulnerability to Anthropogenic Climate Change
A Global-Scale Evaluation of Mammalian Exposure and Vulnerability to Anthropogenic Climate Change Tanya L. Graham A Thesis in The Department of Geography, Planning and Environment Presented in Partial Fulfillment of the Requirements for the Degree of Master of Science (Geography, Urban and Environmental Studies) at Concordia University Montreal, Quebec, Canada March 2018 © Tanya L. Graham, 2018 Abstract A Global-Scale Evaluation of Mammalian Exposure and Vulnerability to Anthropogenic Climate Change Tanya L. Graham There is considerable evidence demonstrating that anthropogenic climate change is impacting species living in the wild. The vulnerability of a given species to such change may be understood as a combination of the magnitude of climate change to which the species is exposed, the sensitivity of the species to changes in climate, and the capacity of the species to adapt to climatic change. I used species distributions and estimates of expected changes in local temperatures per teratonne of carbon emissions to assess the exposure of terrestrial mammal species to human-induced climate change. I evaluated species vulnerability to climate change by combining expected local temperature changes with species conservation status, using the latter as a proxy for species sensitivity and adaptive capacity to climate change. I also performed a global-scale analysis to identify hotspots of mammalian vulnerability to climate change using expected temperature changes, species richness and average species threat level for each km2 across the globe. The average expected change in local annual average temperature for terrestrial mammal species is 1.85 oC/TtC. Highest temperature changes are expected for species living in high northern latitudes, while smaller changes are expected for species living in tropical locations. -

PUBLICATIONS of ROBERT D. BRADLEY 2019 190. Phillips, Caleb D., Jonathan L. Dunnam, Robert C. Dowler, Lisa C. Bradley, Heath Ga
PUBLICATIONS OF ROBERT D. BRADLEY 2019 190. Phillips, Caleb D., Jonathan L. Dunnam, Robert C. Dowler, Lisa C. Bradley, Heath Garner, Kathy McDonald, Burton K. Lim, Marcy A. Revelez, Mariel L. Campbell, Joseph A. Cook, Robert D. Bradley, and the Systematic Collections Committee of the American Society of Mammalogists. Curatorial guidelines and standards of the American Society of Mammalogists for collections of genetic resource. Journal of Mammalogy 100:1690- 1694. 189. Bradley, Robert D. 2019. On being a graduate student of Robert J. Baker: prospects, perils, and philosophies – lessons learned. Pp. 847-860 in From field to laboratory: A memorial volume in honor of Robert J. Baker (R. D. Bradley, H. H. Genoways, D. J. Schmidly, and L. C. Bradley, eds.). Special Publications, Museum of Texas Tech University 71:xi+1-911. 188. Swier, Vicki J., Robert D. Bradley, Frederick F. B. Elder, and Robert J. Baker. 2019. Primitive karyotype for Muroidea: evidence from chromosome paints and fluorescent G- bands. Pp. 629-642 in From field to laboratory: A memorial volume in honor of Robert J. Baker (R. D. Bradley, H. H. Genoways, D. J. Schmidly, and L. C. Bradley, eds.). Special Publications, Museum of Texas Tech University 71:xi+1-911. 187. Keith, Megan S., Roy Neal Platt II, and Robert D. Bradley. 2019. Molecular data indicate that Isthmomys is not aligned with Peromyscus. Pp. 613-628 in From field to laboratory: A memorial volume in honor of Robert J. Baker (R. D. Bradley, H. H. Genoways, D. J. Schmidly, and L. C. Bradley, eds.). Special Publications, Museum of Texas Tech University 71:xi+1-911. -

Chec List Mammals of the San Pedro-Mezquital River Basin, Durango-Nayarit, Mexico
Check List 10(6): 1277–1289, 2014 © 2014 Check List and Authors Chec List ISSN 1809-127X (available at www.biotaxa.org/cl) Journal of species lists and distribution Mammals of the San Pedro-Mezquital River Basin, PECIES S Durango-Nayarit, Mexico OF ISTS Celia López-González *, Abraham Lozano, Diego F. García-Mendoza, and Alí Ituriel Villanueva- L Hernández Instituto Politécnico Nacional, CIIDIR Unidad Durango, Sigma 119, Fraccionamiento 20 de Noviembre II, Durango, Durango 34220, México * Corresponding author. E-mail: [email protected] Abstract: The San Pedro–Mezquital River Basin is located in the southern Sierra Madre Occidental, at the Nearctic– Neotropical transition. The river traverses the Sierra through a canyon that reaches over 1000 m in depth. Based on examination of museum specimens, literature records, and our own collections, we documented the occurrence of 120 species (24.6% of the Mexican terrestrial mammals), 24 endemic to Mexico. Richness was comparable with other megadiverse areas of Mexico, and higher than any other Nearctic–Neotropical transition area, moreover species richness is thelikely canyon. to rise Anthropogenic as survey continues. threats Contrary including to damming expectation, of the distribution river, uncontrolled of mammals cattle across grazing, the and basin pollution not only from reflected domestic the sources,Nearctic–Neotropical call for effective divide, management but a third strategies fauna that to is preserve a mixture one of oftropical, the most temperate biodiverse and areas desert of speciesMexico. was identifiable at DOI: 10.15560/10.6.1277 Introduction inhabitants, INEGI 2005). Additionally, it is one of the Mexico has one of the most diverse mammalian faunas main providers of water for the Marismas Nacionales, in the world (525 species) surpassed only by Indonesia the most widespread mangrove of the Mexican Pacific and Brazil (Ceballos et al. -

Rodent Systematics in an Age of Discovery: Recent Advances and Prospects
applyparastyle "fig//caption/p[1]" parastyle "FigCapt" applyparastyle "fig" parastyle "Figure" Journal of Mammalogy, 100(3):852–871, 2019 DOI:10.1093/jmammal/gyy179 Rodent systematics in an age of discovery: recent advances and prospects Guillermo D’Elía,* Pierre-Henri Fabre, and Enrique P. Lessa Downloaded from https://academic.oup.com/jmammal/article-abstract/100/3/852/5498027 by Tarrant County College user on 26 May 2019 Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Campus Isla Teja s/n, Valdivia 5090000, Chile (GD) Institut des Sciences de l’Evolution (ISEM, UMR 5554 CNRS-UM2-IRD), Université Montpellier, Place E. Bataillon - CC 064 - 34095 Montpellier Cedex 5, France (P-HF) Departamento de Ecología y Evolución, Facultad de Ciencias, Universidad de la República, Iguá 4225, Montevideo 1400, Uruguay (EPL) * Correspondent: [email protected] With almost 2,600 species, Rodentia is the most diverse order of mammals. Here, we provide an overview of changes in our understanding of the systematics of living rodents, including species recognition and delimitation, phylogenetics, and classification, with emphasis on the last three decades. Roughly, this corresponds to the DNA sequencing era of rodent systematics, but the field is undergoing a transition into the genomic era. At least 248 species were newly described in the period 2000–2017, including novelties such as the first living member of Diatomyidae and a murid species without molars (Paucidentomys vermidax), thus highlighting the fact that our understanding of rodent diversity is going through an age of discovery. Mito-nuclear discordance (including that resulting from mitochondrial introgression) has been detected in some of the few taxonomic studies that have assessed variation of two or more unlinked loci. -

Type Localities of Mexican Land Mammals, with Comments on Taxonomy and Nomenclature
Special Publications Museum of Texas Tech University Number xx73 9xx January XXXX 20202010 Type Localities of Mexican Land Mammals, with Comments on Taxonomy and Nomenclature Alfred L. Gardner and José Ramírez-Pulido Front cover: Edward W. Nelson (right) preparing specimens in camp on Mt. Tancítaro, Michoacán. Photograph by Edward A. Goldman, March 1903. Courtesy of Smithsonian Institution Archives, Nelson Goldman files RU7634. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 73 Type Localities of Mexican Land Mammals, with Comments on Taxonomy and Nomenclature Alfred L. Gardner and José Ramírez-Pulido Layout and Design: Lisa Bradley Cover Design: Photo courtesy of Smithsonian Institution Archives, Nelson Goldman files, RU7634 Production Editor: Lisa Bradley Copyright 2020, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (www.depts.ttu.edu/nsrl). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 9 January 2020 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 73 Series Editor: Robert D. -

65 Years of Museum-Based Mammal Research in México: from Taxonomy to Worldwide Information Networks
THERYA, 2021, Vol. 12(1):57-74 DOI:10.12933/therya-21-978 ISSN 2007-3364 65 years of museum-based mammal research in México: from taxonomy to worldwide information networks CELIA LÓPEZ-GONZÁLEZ1*, CYNTHIA ELIZALDE-ARELLANO2, MIGUEL BRIONES-SALAS3, MARIO C. LAVARIEGA3, AND JUAN CARLOS LÓPEZ-VIDAL2 1 Instituto Politécnico Nacional, Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional (CIIDIR) Unidad Du- rango, Calle Sigma 119 Fracc. 20 de Noviembre II, CP. 34220, Durango. Durango, México. Email: [email protected] (CLG). 2 Instituto Politécnico Nacional, Laboratorio de Cordados Terrestres, Departamento de Zoología, Escuela Nacional de Ciencias Biológicas, Carpio y Plan de Ayala s/n, Col. Casco Santo Tomás, CP. 11340. Ciudad de México, México. Email: thiadeno@hotmail. com (CEA), [email protected] (JCLV). 3 Instituto Politécnico Nacional, Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional (CIIDIR) Unidad Oaxaca, Hornos 1003, Colonia Noche Buena, CP. 71230, Santa Cruz Xoxocotlán. Oaxaca, México. Email: [email protected] (MBS), [email protected] (MCL). *Corresponding author Biological collections have become a key tool for biodiversity research. They are repositories of germplasm and data on modified or ex- tinct natural populations, providing valuable information for understanding anthropogenic impacts on the natural world. We appraised the scientific value of the three mammal collections maintained by the Instituto Politécnico Nacional of Mexico (IPN): Escuela Nacional de Ciencias Biológicas (ENCB), CIIDIR Durango (CRD), and CIIDIR Oaxaca (OAXMA). We evaluated their specimen inventory, geographic coverage, and scientific importance for mammalogy. We assessed their physical conditions and provided insights into their future as data sources for unders- tanding natural changes in the 21st century. -

Peromyscus Newsletter Number 43
Number Forty-Three Fall 2008 Cover: Anastasia Island Beach Mouse, Peromyscus polionotus phasma, in a patch of Spartina patens. Photograph by J.B. Miller, Senior Land Resource Planner, St. Johns River Water Management District. 2 Peromyscus Newsletter Number 43 Hello, All! In this latest issue of Peromyscus Newsletter there are several exciting bits of news for you. There is the announcement of a newly created list serve for Peromyscus researchers, and an update on the ESTs and genome sequence which are almost ready to be posted to GenBank, and some really great research in the in the Contributed Accounts section, to name a few. I hope you enjoy them. If you are planning to purchase mice from the Stock Center, please note that there is a new list of user fees and some stocks are no longer available. Also, if you need to place a large order let Janet know as soon as possible by emailing her at [email protected]. And you should definitely check out the article on Janet on page 16. Those of you who don’t know her will be amazed at all she does and those of you who do know her will nod your head in agreement! As I mentioned in the last issue, many people are still not receiving emails from the [email protected] account. This, I believe, is caused by people’s spam filters, so if you know of anyone having this problem please have them check their filters and specify this address as legitimate. I am limited in what I can do from this end.