A CHOP-Regulated Microrna Controls Rhodopsin Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal Rnase P
biomolecules Review Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P Manoj P. Samanta 1,*,†, Stella M. Lai 2,3,†, Charles J. Daniels 3,4 and Venkat Gopalan 2,3,* 1 Systemix Institute, Redmond, WA 98053, USA 2 Department of Chemistry & Biochemistry, The Ohio State University, Columbus, OH 43210, USA; [email protected] 3 Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA; [email protected] 4 Department of Microbiology, The Ohio State University, Columbus, OH 43210, USA * Correspondence: [email protected] (M.P.S.); [email protected] (V.G.); Tel.: +1-425-898-8187 (M.P.S.); +1-614-292-1332 (V.G.) † These authors contributed equally to this work. Academic Editor: Denis Drainas Received: 1 March 2016; Accepted: 8 April 2016; Published: 20 April 2016 Abstract: RNase P, a ribozyme-based ribonucleoprotein (RNP) complex that catalyzes tRNA 51-maturation, is ubiquitous in all domains of life, but the evolution of its protein components (RNase P proteins, RPPs) is not well understood. Archaeal RPPs may provide clues on how the complex evolved from an ancient ribozyme to an RNP with multiple archaeal and eukaryotic (homologous) RPPs, which are unrelated to the single bacterial RPP. Here, we analyzed the sequence and structure of archaeal RPPs from over 600 available genomes. All five RPPs are found in eight archaeal phyla, suggesting that these RPPs arose early in archaeal evolutionary history. The putative ancestral genomic loci of archaeal RPPs include genes encoding several members of ribosome, exosome, and proteasome complexes, which may indicate coevolution/coordinate regulation of RNase P with other core cellular machineries. -

Download on 20
bioRxiv preprint doi: https://doi.org/10.1101/850776; this version posted January 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Intramembrane protease RHBDL4 cleaves oligosaccharyltransferase subunits to target them for ER-associated degradation Julia D. Knopf1, Nina Landscheidt1, Cassandra L. Pegg2, Benjamin L. Schulz2, Nathalie Kühnle1, Chao-Wei Chao1, Simon Huck1 and Marius K. Lemberg1, # 1Centre for Molecular Biology of Heidelberg University (ZMBH), DKFZ-ZMBH Alliance, 69120 Heidelberg, Germany. 2School of Chemistry and Molecular Biosciences, ARC Training Centre for Biopharmaceutical Innovation, The University of Queensland, St Lucia QLD 4072, Australia. #Corresponding author: [email protected] Running title: RHBDL4 triggers ERAD of OST subunits Key words: Rhomboid serine protease, Rhbdd1, ubiquitin-dependent proteolysis, post- translational protein abundance control, N-linked glycosylation. Abbreviations ERAD, ER-associated degradation; OST, oligosacharyltransferase; TM, transmembrane; UIM, ubiquitin-interacting motif. Abstract The Endoplasmic Reticulum (ER)-resident intramembrane rhomboid protease RHBDL4 generates metastable protein fragments and together with the ER-associated degradation (ERAD) machinery provides a clearance mechanism for aberrant and surplus proteins. However, the endogenous substrate spectrum and with that the role of RHBDL4 in physiological ERAD is mainly unknown. Here, we use a substrate trapping approach in combination with quantitative proteomics to identify physiological RHBDL4 substrates. This revealed oligosacharyltransferase (OST) complex subunits such as the catalytic active subunit STT3A as substrates for the RHBDL4-dependent ERAD pathway. RHBDL4-catalyzed cleavage inactivates OST subunits by triggering dislocation into the cytoplasm and subsequent proteasomal degradation. -

Protein Expression Analysis of an in Vitro Murine Model of Prostate Cancer Progression: Towards Identification of High-Potential Therapeutic Targets
Journal of Personalized Medicine Article Protein Expression Analysis of an In Vitro Murine Model of Prostate Cancer Progression: Towards Identification of High-Potential Therapeutic Targets Hisham F. Bahmad 1,2,3 , Wenjing Peng 4, Rui Zhu 4, Farah Ballout 1, Alissar Monzer 1, 1,5 6, , 1, , 4, , Mohamad K. Elajami , Firas Kobeissy * y , Wassim Abou-Kheir * y and Yehia Mechref * y 1 Department of Anatomy, Cell Biology and Physiological Sciences, Faculty of Medicine, American University of Beirut, Beirut 1107-2020, Lebanon; [email protected] (H.F.B.); [email protected] (F.B.); [email protected] (A.M.); [email protected] (M.K.E.) 2 Arkadi M. Rywlin M.D. Department of Pathology and Laboratory Medicine, Mount Sinai Medical Center, Miami Beach, FL 33140, USA 3 Herbert Wertheim College of Medicine, Florida International University, Miami, FL 33199, USA 4 Department of Chemistry and Biochemistry, Texas Tech University, Lubbock, TX 79409, USA; [email protected] (W.P.); [email protected] (R.Z.) 5 Department of Internal Medicine, Mount Sinai Medical Center, Miami Beach, FL 33140, USA 6 Department of Biochemistry and Molecular Genetics, Faculty of Medicine, American University of Beirut, Beirut 1107-2020, Lebanon * Correspondence: [email protected] (F.K.); [email protected] (W.A.-K.); [email protected] (Y.M.); Tel.: +961-1-350000 (ext. 4805) (F.K.); +961-1-350000 (ext. 4778) (W.A.K.); +1-806-834-8246 (Y.M.); Fax: +1-806-742-1289 (Y.M.); 961-1-744464 (W.A.K.) These authors have contributed equally to this work as joint senior authors. -
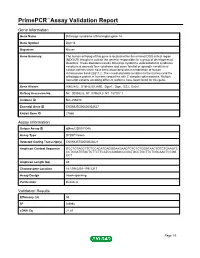
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DiGeorge syndrome critical region gene 14 Gene Symbol Dgcr14 Organism Mouse Gene Summary The human ortholog of this gene is located within the minimal DGS critical region (MDGCR) thought to contain the gene(s) responsible for a group of developmental disorders. These disorders include DiGeorge syndrome velocardiofacial syndrome conotruncal anomaly face syndrome and some familial or sporadic conotruncal cardiac defects which have been associated with microdeletion of human chromosome band 22q11.2. The encoded protein localizes to the nucleus and the orthologous protein in humans co-purifies with C complex spliceosomes. Multiple transcript variants encoding different isoforms have been found for this gene. Gene Aliases AI462402, D16H22S1269E, Dgcr1, Dgsi, ES2, Es2el RefSeq Accession No. NC_000082.6, NT_039624.8, NT_187007.1 UniGene ID Mm.256480 Ensembl Gene ID ENSMUSG00000003527 Entrez Gene ID 27886 Assay Information Unique Assay ID qMmuCID0011048 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000003621 Amplicon Context Sequence GCCTGTAGCTTCTCCACATCAGGGAAGAAGTCTCTCTGGATAACTGTCTGAAGTC CCTCGATGTACTCTTCTTCATCCAGGACCCGCTGCCTGCTTCTCGCAACTCCGG CCT Amplicon Length (bp) 82 Chromosome Location 16:17910201-17911217 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 95 R2 0.9994 cDNA Cq 21.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79.5 gDNA Cq 23.94 Specificity (%) 100 Information to assist with data interpretation is provided -

CRNKL1 Is a Highly Selective Regulator of Intron-Retaining HIV-1 and Cellular Mrnas
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CRNKL1 is a highly selective regulator of intron-retaining HIV-1 and cellular mRNAs 2 3 4 Han Xiao1, Emanuel Wyler2#, Miha Milek2#, Bastian Grewe3, Philipp Kirchner4, Arif Ekici4, Ana Beatriz 5 Oliveira Villela Silva1, Doris Jungnickl1, Markus Landthaler2,5, Armin Ensser1, and Klaus Überla1* 6 7 1 Institute of Clinical and Molecular Virology, University Hospital Erlangen, Friedrich-Alexander 8 Universität Erlangen-Nürnberg, Erlangen, Germany 9 2 Berlin Institute for Medical Systems Biology, Max-Delbrück-Center for Molecular Medicine in the 10 Helmholtz Association, Robert-Rössle-Strasse 10, 13125, Berlin, Germany 11 3 Department of Molecular and Medical Virology, Ruhr-University, Bochum, Germany 12 4 Institute of Human Genetics, University Hospital Erlangen, Friedrich-Alexander Universität Erlangen- 13 Nürnberg, Erlangen, Germany 14 5 IRI Life Sciences, Institute für Biologie, Humboldt Universität zu Berlin, Philippstraße 13, 10115, Berlin, 15 Germany 16 # these two authors contributed equally 17 18 19 *Corresponding author: 20 Prof. Dr. Klaus Überla 21 Institute of Clinical and Molecular Virology, University Hospital Erlangen 22 Friedrich-Alexander Universität Erlangen-Nürnberg 23 Schlossgarten 4, 91054 Erlangen 24 Germany 25 Tel: (+49) 9131-8523563 26 e-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

Análise Correlacional Entre a Expressão Dos Fatores De Splicing E a Ocorrência De Splicing Alternativo Em Tecidos Humanos E De Camundongos
ANÁLISE CORRELACIONAL ENTRE A EXPRESSÃO DOS FATORES DE SPLICING E A OCORRÊNCIA DE SPLICING ALTERNATIVO EM TECIDOS HUMANOS E DE CAMUNDONGOS JULIO CÉSAR NUNES Dissertação apresentada à Fundação Antônio Prudente para a obtenção do título de Mestre em Ciências Área de Concentração: Oncologia Orientador: Dr. Sandro José de Souza São Paulo 2008 Livros Grátis http://www.livrosgratis.com.br Milhares de livros grátis para download. FICHA CATALOGRÁFICA Preparada pela Biblioteca da Fundação Antônio Prudente Nunes, Julio César Análise correlacional entre a expressão dos fatores de splicing e a ocorrência de splicing alternativo em tecidos humanos e de camundongos / Julio César Nunes – São Paulo, 2008. 79p. Dissertação (Mestrado) - Fundação Antônio Prudente. Curso de Pós-Graduação em Ciências - Área de concentração: Oncologia. Orientador: Sandro José Souza Descritores: 1. SPLICING ALTERNATIVO 2. BIOLOGIA MOLECULAR COMPUTACIONAL 3. CÂNCER 4. GENOMICA. AGRADECIMENTOS Agradeço à FAPESP e CAPES pela bolsa de Mestrado. Ao Sandro José de Souza agradeço toda orientação e conhecimento oferecido. Meus especiais agradecimentos ao Pedro Alexandre Favoretto Galante que dedicou atenção a minha formação no processo de Pós-Graduação na Fundação Antônio Prudente, bem como pela sua oficiosa co-orientação ao projeto de pesquisa. À grande família e amigos pela dedicação e incentivo a minha formação acadêmica. À Fundação Antônio Prudente, Hospital do Câncer e Instituto Ludwig de Pesquisa sobre o Câncer dedico os meus nobres agradecimentos finais. RESUMO Nunes JC. Análise correlacional entre a expressão dos fatores de splicing e a ocorrência de splicing alternativo em tecidos humanos e de camundongos. São Paulo; 2007. [Dissertacão de Mestrado - Fundação Antônio Prudente] Splicing alternativo desempenha uma significante função no aumento da complexidade genômica, produzindo um extenso número de mRNA e isoformas protéicas. -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

DGCR6 at the Proximal Part of the Digeorge Critical Region Is Involved in Conotruncal Heart Defects
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Institutional Repository : the EHIME area OPEN Citation: Human Genome Variation (2015) 2, 15004; doi:10.1038/hgv.2015.4 © 2015 The Japan Society of Human Genetics All rights reserved 2054-345X/15 www.nature.com/hgv ARTICLE DGCR6 at the proximal part of the DiGeorge critical region is involved in conotruncal heart defects Wenming Gao1, Takashi Higaki1, Minenori Eguchi-Ishimae1, Hidehiko Iwabuki1, Zhouying Wu1, Eiichi Yamamoto2, Hidemi Takata1, Masaaki Ohta1, Issei Imoto3, Eiichi Ishii1 and Mariko Eguchi1 Cardiac anomaly is one of the hallmarks of DiGeorge syndrome (DGS), observed in approximately 80% of patients. It often shows a characteristic morphology, termed as conotruncal heart defects. In many cases showing only the conotruncal heart defect, deletion of 22q11.2 region cannot be detected by fluorescence in situ hybridization (FISH), which is used to detect deletion in DGS. We investigated the presence of genomic aberrations in six patients with congenital conotruncal heart defects, who show no deletion at 22q11.2 in an initial screening by FISH. In these patients, no abnormalities were identified in the coding region of the TBX1 gene, one of the key genes responsible for the phenotype of DGS. However, when copy number alteration was analyzed by high-resolution array analysis, a small deletion or duplication in the proximal end of DiGeorge critical region was detected in two patients. The affected region contains the DGCR6 and PRODH genes. DGCR6 has been reported to affect the expression of the TBX1 gene. Our results suggest that altered dosage of gene(s) other than TBX1, possibly DGCR6, may also be responsible for the development of conotruncal heart defects observed in patients with DGS and, in particular, in those with stand-alone conotruncal heart defects. -

Nuclear PTEN Safeguards Pre-Mrna Splicing to Link Golgi Apparatus for Its Tumor Suppressive Role
ARTICLE DOI: 10.1038/s41467-018-04760-1 OPEN Nuclear PTEN safeguards pre-mRNA splicing to link Golgi apparatus for its tumor suppressive role Shao-Ming Shen1, Yan Ji2, Cheng Zhang1, Shuang-Shu Dong2, Shuo Yang1, Zhong Xiong1, Meng-Kai Ge1, Yun Yu1, Li Xia1, Meng Guo1, Jin-Ke Cheng3, Jun-Ling Liu1,3, Jian-Xiu Yu1,3 & Guo-Qiang Chen1 Dysregulation of pre-mRNA alternative splicing (AS) is closely associated with cancers. However, the relationships between the AS and classic oncogenes/tumor suppressors are 1234567890():,; largely unknown. Here we show that the deletion of tumor suppressor PTEN alters pre-mRNA splicing in a phosphatase-independent manner, and identify 262 PTEN-regulated AS events in 293T cells by RNA sequencing, which are associated with significant worse outcome of cancer patients. Based on these findings, we report that nuclear PTEN interacts with the splicing machinery, spliceosome, to regulate its assembly and pre-mRNA splicing. We also identify a new exon 2b in GOLGA2 transcript and the exon exclusion contributes to PTEN knockdown-induced tumorigenesis by promoting dramatic Golgi extension and secretion, and PTEN depletion significantly sensitizes cancer cells to secretion inhibitors brefeldin A and golgicide A. Our results suggest that Golgi secretion inhibitors alone or in combination with PI3K/Akt kinase inhibitors may be therapeutically useful for PTEN-deficient cancers. 1 Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China. 2 Institute of Health Sciences, Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences and SJTU-SM, Shanghai 200025, China. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing