Phylogenetic Systematics and Evolution of the Spider Infraorder Mygalomorphae Using Genomic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biogeography of the Caribbean Cyrtognatha Spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J
www.nature.com/scientificreports OPEN Biogeography of the Caribbean Cyrtognatha spiders Klemen Čandek1,6,7, Ingi Agnarsson2,4, Greta J. Binford3 & Matjaž Kuntner 1,4,5,6 Island systems provide excellent arenas to test evolutionary hypotheses pertaining to gene fow and Received: 23 July 2018 diversifcation of dispersal-limited organisms. Here we focus on an orbweaver spider genus Cyrtognatha Accepted: 1 November 2018 (Tetragnathidae) from the Caribbean, with the aims to reconstruct its evolutionary history, examine Published: xx xx xxxx its biogeographic history in the archipelago, and to estimate the timing and route of Caribbean colonization. Specifcally, we test if Cyrtognatha biogeographic history is consistent with an ancient vicariant scenario (the GAARlandia landbridge hypothesis) or overwater dispersal. We reconstructed a species level phylogeny based on one mitochondrial (COI) and one nuclear (28S) marker. We then used this topology to constrain a time-calibrated mtDNA phylogeny, for subsequent biogeographical analyses in BioGeoBEARS of over 100 originally sampled Cyrtognatha individuals, using models with and without a founder event parameter. Our results suggest a radiation of Caribbean Cyrtognatha, containing 11 to 14 species that are exclusively single island endemics. Although biogeographic reconstructions cannot refute a vicariant origin of the Caribbean clade, possibly an artifact of sparse outgroup availability, they indicate timing of colonization that is much too recent for GAARlandia to have played a role. Instead, an overwater colonization to the Caribbean in mid-Miocene better explains the data. From Hispaniola, Cyrtognatha subsequently dispersed to, and diversifed on, the other islands of the Greater, and Lesser Antilles. Within the constraints of our island system and data, a model that omits the founder event parameter from biogeographic analysis is less suitable than the equivalent model with a founder event. -

Cryptic Species Delimitation in the Southern Appalachian Antrodiaetus
Cryptic species delimitation in the southern Appalachian Antrodiaetus unicolor (Araneae: Antrodiaetidae) species complex using a 3RAD approach Lacie Newton1, James Starrett1, Brent Hendrixson2, Shahan Derkarabetian3, and Jason Bond4 1University of California Davis 2Millsaps College 3Harvard University 4UC Davis May 5, 2020 Abstract Although species delimitation can be highly contentious, the development of reliable methods to accurately ascertain species boundaries is an imperative step in cataloguing and describing Earth's quickly disappearing biodiversity. Spider species delimi- tation remains largely based on morphological characters; however, many mygalomorph spider populations are morphologically indistinguishable from each other yet have considerable molecular divergence. The focus of our study, Antrodiaetus unicolor species complex which contains two sympatric species, exhibits this pattern of relative morphological stasis with considerable genetic divergence across its distribution. A past study using two molecular markers, COI and 28S, revealed that A. unicolor is paraphyletic with respect to A. microunicolor. To better investigate species boundaries in the complex, we implement the cohesion species concept and employ multiple lines of evidence for testing genetic exchangeability and ecological interchange- ability. Our integrative approach includes extensively sampling homologous loci across the genome using a RADseq approach (3RAD), assessing population structure across their geographic range using multiple genetic clustering analyses that include STRUCTURE, PCA, and a recently developed unsupervised machine learning approach (Variational Autoencoder). We eval- uate ecological similarity by using large-scale ecological data for niche-based distribution modeling. Based on our analyses, we conclude that this complex has at least one additional species as well as confirm species delimitations based on previous less comprehensive approaches. -

Five New Species of the Open-Holed Trapdoor Spider Genus Aname
RECORDS OF THE WESTERN AUSTRALIAN MUSEUM 35 010–038 (2020) DOI: 10.18195/issn.0312-3162.35.2020.010-038 Five new species of the open-holed trapdoor spider genus Aname (Araneae: Mygalomorphae: Anamidae) from Western Australia, with a revised generic placement for Aname armigera Mark S. Harvey1,2, Karl Gruber2, Mia J. Hillyer1 and Joel A. Huey1,2,3,4 1 Collections and Research, Western Australian Museum, 49 Kew Street, Welshpool, Western Australia 6106, Australia. 2 School of Biological Sciences, University of Western Australia, Crawley, Western Australia 6009, Australia. 3 Adjunct, School of Natural Sciences, Edith Cowan University, Joondalup, Western Australia 6027, Australia. 4 Present address: Biologic Environmental Survey, East Perth, Western Australia 6004, Australia. Corresponding author: [email protected] ABSTRACT – The open-holed trapdoor spider genus Aname L. Koch, 1873 is widely distributed throughout Australia, and currently contains 44 named species. Using a combination of morphological and molecular data, we describe fve new species from the Wheatbelt, Mid-west and Goldfelds regions of Western Australia: A. exulans sp. nov., A. lillianae sp. nov., A. mccleeryorum sp. nov., A. phillipae sp. nov. and A. simoneae sp. nov. The female holotype of Aname armigera Rainbow and Pulleine, 1918 from near Mullewa was examined and found to belong to the genus Proshermacha Simon forming the new combination P. armigera (Rainbow and Pulleine, 1918), comb. nov. KEYWORDS: taxonomy, systematics, molecular phylogenetics urn:lsid:zoobank.org:pub:98828964-6150-465D-B5AE-1480DA0D454E INTRODUCTION Castalanelli, Framenau, Huey and Harvey, 2020, a The open-holed trapdoor spider genus Aname species from Western Australia recently described in L. -

Final Project Completion Report
CEPF SMALL GRANT FINAL PROJECT COMPLETION REPORT Organization Legal Name: - Tarantula (Araneae: Theraphosidae) spider diversity, distribution and habitat-use: A study on Protected Area adequacy and Project Title: conservation planning at a landscape level in the Western Ghats of Uttara Kannada district, Karnataka Date of Report: 18 August 2011 Dr. Manju Siliwal Wildlife Information Liaison Development Society Report Author and Contact 9-A, Lal Bahadur Colony, Near Bharathi Colony Information Peelamedu Coimbatore 641004 Tamil Nadu, India CEPF Region: The Western Ghats Region (Sahyadri-Konkan and Malnad-Kodugu Corridors). 2. Strategic Direction: To improve the conservation of globally threatened species of the Western Ghats through systematic conservation planning and action. The present project aimed to improve the conservation status of two globally threatened (Molur et al. 2008b, Siliwal et al., 2008b) ground dwelling theraphosid species, Thrigmopoeus insignis and T. truculentus endemic to the Western Ghats through systematic conservation planning and action. Investment Priority 2.1 Monitor and assess the conservation status of globally threatened species with an emphasis on lesser-known organisms such as reptiles and fish. The present project was focused on an ignored or lesser-known group of spiders called Tarantulas/ Theraphosid spiders and provided valuable information on population status and potential conservation sites in Uttara Kannada district, which will help in future monitoring and assessment of conservation status of the two globally threatened theraphosid species T. insignis and Near Threatened T. truculentus. Investment Priority 2.3. Evaluate the existing protected area network for adequate globally threatened species representation and assess effectiveness of protected area types in biodiversity conservation. -

Miranda ZA 2018.Pdf
Zoologischer Anzeiger 273 (2018) 33–55 Contents lists available at ScienceDirect Zoologischer Anzeiger jou rnal homepage: www.elsevier.com/locate/jcz Review of Trichodamon Mello-Leitão 1935 and phylogenetic ଝ placement of the genus in Phrynichidae (Arachnida, Amblypygi) a,b,c,∗ a Gustavo Silva de Miranda , Adriano Brilhante Kury , a,d Alessandro Ponce de Leão Giupponi a Laboratório de Aracnologia, Museu Nacional do Rio de Janeiro, Universidade Federal do Rio de Janeiro, Quinta da Boa Vista s/n, São Cristóvão, Rio de Janeiro-RJ, CEP 20940-040, Brazil b Entomology Department, National Museum of Natural History, Smithsonian Institution, 10th St. & Constitution Ave NW, Washington, DC, 20560, USA c Center for Macroecology, Evolution and Climate, Natural History Museum of Denmark (Zoological Museum), University of Copenhagen, Universitetsparken 15, 2100, Copenhagen, Denmark d Servic¸ o de Referência Nacional em Vetores das Riquetsioses (LIRN), Colec¸ ão de Artrópodes Vetores Ápteros de Importância em Saúde das Comunidades (CAVAISC), IOC-FIOCRUZ, Manguinhos, 21040360, Rio de Janeiro, RJ, Brazil a r t i c l e i n f o a b s t r a c t Article history: Amblypygi Thorell, 1883 has five families, of which Phrynichidae is one of the most diverse and with a Received 18 October 2017 wide geographic distribution. The genera of this family inhabit mostly Africa, India and Southeast Asia, Received in revised form 27 February 2018 with one genus known from the Neotropics, Trichodamon Mello-Leitão, 1935. Trichodamon has two valid Accepted 28 February 2018 species, T. princeps Mello-Leitão, 1935 and T. froesi Mello-Leitão, 1940 which are found in Brazil, in the Available online 10 March 2018 states of Bahia, Goiás, Minas Gerais and Rio Grande do Norte. -

The Case of Embrik Strand (Arachnida: Araneae) 22-29 Arachnologische Mitteilungen / Arachnology Letters 59: 22-29 Karlsruhe, April 2020
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Arachnologische Mitteilungen Jahr/Year: 2020 Band/Volume: 59 Autor(en)/Author(s): Nentwig Wolfgang, Blick Theo, Gloor Daniel, Jäger Peter, Kropf Christian Artikel/Article: How to deal with destroyed type material? The case of Embrik Strand (Arachnida: Araneae) 22-29 Arachnologische Mitteilungen / Arachnology Letters 59: 22-29 Karlsruhe, April 2020 How to deal with destroyed type material? The case of Embrik Strand (Arachnida: Araneae) Wolfgang Nentwig, Theo Blick, Daniel Gloor, Peter Jäger & Christian Kropf doi: 10.30963/aramit5904 Abstract. When the museums of Lübeck, Stuttgart, Tübingen and partly of Wiesbaden were destroyed during World War II between 1942 and 1945, also all or parts of their type material were destroyed, among them types from spider species described by Embrik Strand bet- ween 1906 and 1917. He did not illustrate type material from 181 species and one subspecies and described them only in an insufficient manner. These species were never recollected during more than 110 years and no additional taxonomically relevant information was published in the arachnological literature. It is impossible to recognize them, so we declare these 181 species here as nomina dubia. Four of these species belong to monotypic genera, two of them to a ditypic genus described by Strand in the context of the mentioned species descriptions. Consequently, without including valid species, the five genera Carteroniella Strand, 1907, Eurypelmella Strand, 1907, Theumella Strand, 1906, Thianella Strand, 1907 and Tmeticides Strand, 1907 are here also declared as nomina dubia. Palystes modificus minor Strand, 1906 is a junior synonym of P. -

Eg the Short Range-Endemics of the Pilbara Bioregion
Appendix 3 Supporting Technical Studies Earl Grey Lithium Project SRE and Subterranean Fauna Desktop Assessment Prepared for: Covalent Lithium January 2019 Final Report May 2017 Earl Grey SRE & Subterranean Fauna Kidman Resources Ltd Earl Grey Lithium Project SRE and Subterranean Fauna Desktop Assessment Bennelongia Pty Ltd 5 Bishop Street Jolimont WA 6014 P: (08) 9285 8722 F: (08) 9285 8811 E: [email protected] ABN: 55 124 110 167 Report Number: 298 Report Version Prepared by Reviewed by Submitted to Client Method Date Draft Anton Mittra Stuart Halse Email 31 May 2017 Final Stuart Halse Email 24 November 17 Final V2 Anton Mittra Email 14 January 2019 BEC_Mt Holland_SRE_final_V2_10i2019.docx This document has been prepared to the requirements of the Client and is for the use by the Client, its agents, and Bennelongia Environmental Consultants. Copyright and any other Intellectual Property associated with the document belongs to Bennelongia Environmental Consultants and may not be reproduced without written permission of the Client or Bennelongia. No liability or responsibility is accepted in respect of any use by a third party or for purposes other than for which the document was commissioned. Bennelongia has not attempted to verify the accuracy and completeness of information supplied by the Client. © Copyright 2015 Bennelongia Pty Ltd. i Earl Grey SRE & Subterranean Fauna Kidman Resources Ltd EXECUTIVE SUMMARY Covalent Lithium proposes to mine lithium at the Earl Grey deposit (the Proposal) approximately 100 km southeast of Southern Cross in Western Australia. This desktop review examines the likelihood that short-range endemic (SRE) invertebrates and listed terrestrial invertebrate species occur in the Proposal area and whether these species are likely to be impacted by proposed development. -

Species Are Hypotheses: Avoid Connectivity Assessments Based on Pillars of Sand Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D
Species are hypotheses: avoid connectivity assessments based on pillars of sand Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D. Aurelle, Magalie Castelin, Anne Chenuil, Christophe Destombe, Didier Forcioli, Myriam Valero, et al. To cite this version: Eric Pante, Nicolas Puillandre, Amélia Viricel, Sophie Arnaud-Haond, D. Aurelle, et al.. Species are hypotheses: avoid connectivity assessments based on pillars of sand. Molecular Ecology, Wiley, 2015, 24 (3), pp.525-544. hal-02002440 HAL Id: hal-02002440 https://hal.archives-ouvertes.fr/hal-02002440 Submitted on 31 Jan 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Molecular Ecology Species are hypotheses : avoid basing connectivity assessments on pillars of sand. Journal:For Molecular Review Ecology Only Manuscript ID: Draft Manuscript Type: Invited Reviews and Syntheses Date Submitted by the Author: n/a Complete List of Authors: Pante, Eric; UMR 7266 CNRS - Université de La Rochelle, Puillandre, Nicolas; MNHN, Systematique & Evolution Viricel, Amélia; UMR 7266 CNRS - -

Species Delimitation and Phylogeography of Aphonopelma Hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic Diversity in North American Tarantulas
Species Delimitation and Phylogeography of Aphonopelma hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic Diversity in North American Tarantulas Chris A. Hamilton1*, Daniel R. Formanowicz2, Jason E. Bond1 1 Auburn University Museum of Natural History and Department of Biological Sciences, Auburn University, Auburn, Alabama, United States of America, 2 Department of Biology, The University of Texas at Arlington, Arlington, Texas, United States of America Abstract Background: The primary objective of this study is to reconstruct the phylogeny of the hentzi species group and sister species in the North American tarantula genus, Aphonopelma, using a set of mitochondrial DNA markers that include the animal ‘‘barcoding gene’’. An mtDNA genealogy is used to consider questions regarding species boundary delimitation and to evaluate timing of divergence to infer historical biogeographic events that played a role in shaping the present-day diversity and distribution. We aimed to identify potential refugial locations, directionality of range expansion, and test whether A. hentzi post-glacial expansion fit a predicted time frame. Methods and Findings: A Bayesian phylogenetic approach was used to analyze a 2051 base pair (bp) mtDNA data matrix comprising aligned fragments of the gene regions CO1 (1165 bp) and ND1-16S (886 bp). Multiple species delimitation techniques (DNA tree-based methods, a ‘‘barcode gap’’ using percent of pairwise sequence divergence (uncorrected p- distances), and the GMYC method) consistently recognized a number of divergent and genealogically exclusive groups. Conclusions: The use of numerous species delimitation methods, in concert, provide an effective approach to dissecting species boundaries in this spider group; as well they seem to provide strong evidence for a number of nominal, previously undiscovered, and cryptic species. -

Raven, RJ 1984. a Revision of the Aname Maculata Species Group
Raven,R. J. 1984.A revision of the Anamemaculata species group(Araneae, Dipluridae) with notes on biogeography.J. ArachnoL,12:177-193. A REVISION OF THE ANAME MA CULA TA SPECIES GROUP (DIPLURIDAE, ARANEAE) WITH NOTES ON BIOGEOGRAPHY Robert1 J. Raven Queensland Museum Gregory Terrace, Fortitude Valley, Queensland ABSTRACT Thespecies of the Anamem¢culata (Hogg) species-group (previously Chenistonia) are revised. The type species, Chenistonlamaculara Hogg, and C trevallynia Hickmanare diagnosed.Five new species: A. caeruleomontana,A. earrhwatchorum,A. hiekmani,A. montanaand A. tropica, are described. Asmost of these species possess a serrula, absent in manyother species of Aname,the groupis of phylo~eneticsignificance. Becausethe groupoccurs in discontinuousmontane talnforests fromnorthern Queensland to Tasmania,it is also of biogeographicinterest. INTRODUCTION The Anamemaculata species group includes someof those species previously included in Chenistonia which Raven (1981) considered monophyletic. Although the species have revised lack a synapomorphythey remain a coherent taxonomicunit. Males of all species have a moderately short embolus on the palp and the first metatarsus is not usually as incrassate as that of the A. pallida species group. Twopreviously described species, Anamemaculata (Hogg) and Anametrevallynia (Hickman), are included in the group present. The male of "Chenistonia tepperi’ Hogg[presumably that described by Rainbow and PuUeine(1918) as Chenistonia major Hoggand placed by Main (1972) in Stanwellia] is being revised by Mainas part of the very complex’Chenistonia tepperi’ species group. MATERIALS AND METHODS All drawings were made with a camera-lucida. Spermathecae were drawn after being cleared in lactic acid. All measurementsare in millimetres except eye measurements which are in ocular micrometerunits. -
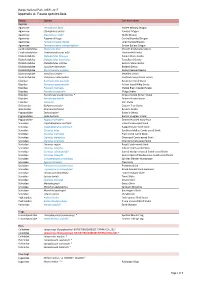
Report-Mungo National Park-Appendix A
Mungo National Park, NSW, 2017 Appendix A: Fauna species lists Family Species Common name Reptiles Agamidae Ctenophorus fordi Mallee Military Dragon Agamidae Ctenophorus pictus Painted Dragon Agamidae Diporiphora nobbi Nobbi Dragon Agamidae Pogona vitticeps Central Bearded Dragon Agamidae Tympanocryptis lineata Lined Earless Dragon Agamidae Tympanocryptis tetraporophora Eyrean Earless Dragon Carphodactylidae Nephrurus levis Smooth Knob-tailed Gecko Carphodactylidae Underwoodisaurus milii Thick-tailed Gecko Diplodactylidae Diplodactylus furcosus Ranges Stone Gecko Diplodactylidae Diplodactylus tessellatus Tessellated Gecko Diplodactylidae Diplodactylus vittatus Eastern Stone Gecko Diplodactylidae Lucasium damaeum Beaded Gecko Diplodactylidae Rhynchoedura ormsbyi Eastern Beaked Gecko Diplodactylidae Strophurus elderi ~ Jewelled Gecko Diplodactylidae Strophurus intermedius Southern Spiny-tailed Gecko Elapidae Brachyurophis australis Australian Coral Snake Elapidae Demansia psammophis Yellow-faced Whip Snake Elapidae Parasuta nigriceps Mallee Black-headed Snake Elapidae Pseudechis australis Mulga Snake Elapidae Pseudonaja aspidorhyncha * Strap-snouted Brown Snake Elapidae Pseudonaja textilis Eastern Brown Snake Elapidae Suta suta Curl Snake Gekkonidae Gehyra versicolor Eastern Tree Gecko Gekkonidae Heteronotia binoei Bynoe's Gecko Pygopodidae Delma butleri Butler's Delma Pygopodidae Lialis burtonis Burton's Legless Lizard Pygopodidae Pygopus schraderi Eastern Hooded Scaly-foot Scincidae Cryptoblepharus australis Inland Snake-eyed Skink Scincidae -

Rossi Gf Me Rcla Par.Pdf (1.346Mb)
RESSALVA Atendendo solicitação da autora, o texto completo desta dissertação será disponibilizado somente a partir de 28/02/2021. UNIVERSIDADE ESTADUAL PAULISTA “JÚLIO DE MESQUITA FILHO” Instituto de Biociências – Rio Claro Departamento de Zoologia Giullia de Freitas Rossi Taxonomia e biogeografia de aranhas cavernícolas da infraordem Mygalomorphae RIO CLARO – SP Abril/2019 Giullia de Freitas Rossi Taxonomia e biogeografia de aranhas cavernícolas da infraordem Mygalomorphae Dissertação apresentada ao Departamento de Zoologia do Instituto de Biociências de Rio Claro, como requisito para conclusão de Mestrado do Programa de Pós-Graduação em Zoologia. Orientador: Prof. Dr. José Paulo Leite Guadanucci RIO CLARO – SP Abril/2019 Rossi, Giullia de Freitas R832t Taxonomia e biogeografia de aranhas cavernícolas da infraordem Mygalomorphae / Giullia de Freitas Rossi. -- Rio Claro, 2019 348 f. : il., tabs., fotos, mapas Dissertação (mestrado) - Universidade Estadual Paulista (Unesp), Instituto de Biociências, Rio Claro Orientador: José Paulo Leite Guadanucci 1. Aracnídeo. 2. Ordem Araneae. 3. Sistemática. I. Título. Sistema de geração automática de fichas catalográficas da Unesp. Biblioteca do Instituto de Biociências, Rio Claro. Dados fornecidos pelo autor(a). Essa ficha não pode ser modificada. Dedico este trabalho à minha família. AGRADECIMENTOS Agradeço ao meus pais, Érica e José Leandro, ao meu irmão Pedro, minha tia Jerusa e minha avó Beth pelo apoio emocional não só nesses dois anos de mestrado, mas durante toda a minha vida. À José Paulo Leite Guadanucci, que aceitou ser meu orientador, confiou em mim e ensinou tudo o que sei sobre Mygalomorphae. Ao meu grande amigo Roberto Marono, pelos anos de estágio e companheirismo na UNESP Bauru, onde me ensinou sobre aranhas, e ao incentivo em ir adiante.