Synsepalum (A.DC.) Daniell and Englerophytum K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Renata Gabriela Vila Nova De Lima Filogenia E Distribuição
RENATA GABRIELA VILA NOVA DE LIMA FILOGENIA E DISTRIBUIÇÃO GEOGRÁFICA DE CHRYSOPHYLLUM L. COM ÊNFASE NA SEÇÃO VILLOCUSPIS A. DC. (SAPOTACEAE) RECIFE 2019 RENATA GABRIELA VILA NOVA DE LIMA FILOGENIA E DISTRIBUIÇÃO GEOGRÁFICA DE CHRYSOPHYLLUM L. COM ÊNFASE NA SEÇÃO VILLOCUSPIS A. DC. (SAPOTACEAE) Dissertação apresentada ao Programa de Pós-graduação em Botânica da Universidade Federal Rural de Pernambuco (UFRPE), como requisito para a obtenção do título de Mestre em Botânica. Orientadora: Carmen Silvia Zickel Coorientador: André Olmos Simões Coorientadora: Liliane Ferreira Lima RECIFE 2019 Dados Internacionais de Catalogação na Publicação (CIP) Sistema Integrado de Bibliotecas da UFRPE Biblioteca Central, Recife-PE, Brasil L732f Lima, Renata Gabriela Vila Nova de Filogenia e distribuição geográfica de Chrysophyllum L. com ênfase na seção Villocuspis A. DC. (Sapotaceae) / Renata Gabriela Vila Nova de Lima. – 2019. 98 f. : il. Orientadora: Carmen Silvia Zickel. Coorientadores: André Olmos Simões e Liliane Ferreira Lima. Dissertação (Mestrado) – Universidade Federal Rural de Pernambuco, Programa de Pós-Graduação em Botânica, Recife, BR-PE, 2019. Inclui referências e anexo(s). 1. Mata Atlântica 2. Filogenia 3. Plantas florestais 4. Sapotaceae I. Zickel, Carmen Silvia, orient. II. Simões, André Olmos, coorient. III. Lima, Liliane Ferreira, coorient. IV. Título CDD 581 ii RENATA GABRIELA VILA NOVA DE LIMA Filogenia e distribuição geográfica de Chrysophyllum L. com ênfase na seção Villocuspis A. DC. (Sapotaceae Juss.) Dissertação apresentada e -

Finding Fruit of This Species Proved Difficult and I Have Only Seen One
Bothalia 23,2(1993) 235 posed by erosion of the soil above it and was probably not quite fully developed. It contained eight relatively large seeds (some dissected ovaries contained up to 10 ovules) and was extremely smelly when opened. In fact, when damaged all parts of the plant give off the unpleas ant foetid odour characteristic of Kedrostis. Meeuse men tions that C. humifructus is dispersed by antbears. I have, though, noticed no particular concentrations of K. psammophila around the groundsquirrel warrens which are common in the sandier parts of Namaqualand and its seeds may be dispersed by moles. REFERENCES DE WET, B.C., ARCHER, R., FISH, L., GERMISHUIZEN, G., HER MAN, P.P., JORDAAN, M., PEROLD, S.M., REID, C., VAN ROOY, J., WTiLMAN. W.G. & GLEN, H.F. 1991. New taxa, new records and name changes for southern African plants. Bothalia 21: 211. GIBBS RUSSELL, G.E.. WELMAN, W.G.. RETIEF. E., IMMELMAN, K.L., GERMISHUIZEN, G.. PIENAAR, B.J., VAN WYK, M. & FIGURE 6.—Distribution of Kedrostispsammophila. NICHOLAS, A. 1987. List of species of southern African plants. Memoirs of the Botanical Sur\'e\' of South Africa No. 56:201 -203. gneissic sand (much more gravelly than the coastal sands) MEEUSE, A.D.J. 1962. The Cucurbitaceae of southern Africa. Bothalia 8: 1-111. higher up on the southern side of the Khamiesberge as far east as Kliprand. P. BRUYNS* Finding fruit of this species proved difficult and I have * Bolus Herbarium, Univ. of Cape Town, Private Bag, Rondebosch 7700. only seen one (Bruyns 5174). This had been partially ex MS. -
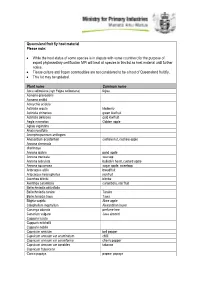
MPI Queensland Fruit Fly Host Material List
Queensland fruit fly host material Please note: While the host status of some species is in dispute with some countries; for the purpose of export phytosanitary certification MPI will treat all species in this list as host material until further notice. Tissue culture and frozen commodities are not considered to be a host of Queensland fruit fly. This list may be updated. Plant name Common name Acca sellowiana (syn Feijoa sellowiana) feijoa Acmena graveolens Acmena smithii Acroychia acidula Actinidia arguta kiwiberry Actinidia chinensis green kiwifruit Actinidia deliciosa gold kiwifruit Aegle marmelos Golden apple Aglaia sapindina Alyxia ruscifolia Amorphospermum antilogum Anacardium occidentale cashew nut, cashew apple Annona cherimola cherimoya Annona glabra pond apple Annona muricata soursop Annona reticulata bullock's heart, custard apple Annona squamosa sugar apple, sweetsop Artocarpus altilis breadfruit Artocarpus heterophyllus jackfruit Averrhoa bilimbi blimbe Averrhoa carambola carambola, star fruit Beilschmiedia obtusifolia Beilschmiedia taraire Taraire Beilschmiedia tawa Tawa Blighia sapida Akee apple Calophyllum inophyllum Alexandrian laurel Cananga odorata perfume tree Canarium vulgare Java almond Capparis lucida Capparis mitchellii Capparis nobilis Capsicum annuum bell pepper Capsicum annuum var acuminatum chilli Capsicum annuum var cerasiforme cherry pepper Capsicum annuum var conoides tabasco Capsicum frutescens Carica papaya papaw, papaya Carica pentagona babaco Carissa ovata Casimiroa edulis white sapote Cassine australia Castanospora alphandii Chrysophyllum cainito caimito, star apple Cissus sp Citrullus lanatus Watermelon Citrus spp. Citrus aurantiifolia West indian lime Citrus aurantium Seville orange Citrus grandis (syn maxima) pummelo Citrus hystrix kaffir lime Citrus jambhiri rough lemon Citrus latifolia Tahatian lime Citrus limetta sweet lemon tree Citrus limon lemon Citrus maxima (syn grandis) pomelo, shaddock Citrus medica citron Citrus meyeri Meyer lemon Citrus reticulata mandarin Citrus reticulata var. -

The Potential of Agroforestry in the Conservation of High Value Indigenous Trees: a Case Study of Umzimvubu District, Eastern Cape
THE POTENTIAL OF AGROFORESTRY IN THE CONSERVATION OF HIGH VALUE INDIGENOUS TREES: A CASE STUDY OF UMZIMVUBU DISTRICT, EASTERN CAPE. SUPERVISOR: PROF. MIKE J. LAWES (FOREST BIODIVERSITY PROGRAMME) MICHAEL O. MUKOLWE MASTERS PROGRAMME IN ENVIRONMENT AND DEVELOPMENT SCHOOL OF ENVIRONMENT AND DEVELOPMENT UNIVERSITY OF NATAL PIETERMARITZBURG JULY 1999 This project was carried out within the Forest Biodiversity Programme School of Botany and Zoology University of Natal, Pietermaritzburg BiODiVERSiTY PROGRAMME UNIVERSITY OF NATAL DEDICATION This work dedicated to Messrs. Toshihiro Shima and Seiichi Mishima, and to my beloved wife Anne Florence Asiko and children Marion Akinyi, Fabian Omondi and Stephen Ochieng "Stevo". 11 ABSTRACT South Africa is not well endowed with indigenous forests which are now known to be degraded and declining at unknown rates. This constitutes a direct threat to quality oflife ofthe resource-poor rural households who directly depend on them and to ecological integrity. It is also recognised that the declining tree resources, particularly the high value indigenous tree species, are increasingly threatened by a number ofgrowing subsistence demands. This emphasised the need to cultivate and conserve high-value tree species such as Englerophytum natalense, Ptaeroxylon obliquum and Millettia grandis on-farm in Umzimvubu District. Agroforestry is recognised as a viable option for optimising land productivity, reducing pressure on the indigenous forests, ensuring a sustainable supply ofdesired tree products and services and improving the quality oflife ofthe resource-poor rural households. This Thesis examines whether agroforestry in Umzimvubu District and similar areas ofSouth Africa has the potential for addressing these needs. It recognises that for successful initiation, implementation and adoption, agroforestry should be considered at two levels, namely, household and institutional. -

Red Data List Special Edition
Newsletter of the Southern African Botanical Diversity Network Volume 6 No. 3 ISSN 1027-4286 November 2001 Invasive Alien Plants Part 2 Southern Mozambique Expedition Living Plant Collections: Lowveld, Mozambique, Namibia REDSABONET NewsDATA Vol. 6 No. 3 November LIST 2001 SPECIAL EDITION153 c o n t e n t s Red Data List Features Special 157 Profile: Ezekeil Kwembeya ON OUR COVER: 158 Profile: Anthony Mapaura Ferraria schaeferi, a vulnerable 162 Red Data Lists in Southern Namibian near-endemic. 159 Tribute to Paseka Mafa (Photo: G. Owen-Smith) Africa: Past, Present, and Future 190 Proceedings of the GTI Cover Stories 169 Plant Red Data Books and Africa Regional Workshop the National Botanical 195 Herbarium Managers’ 162 Red Data List Special Institute Course 192 Invasive Alien Plants in 170 Mozambique RDL 199 11th SSC Workshop Southern Africa 209 Further Notes on South 196 Announcing the Southern 173 Gauteng Red Data Plant Africa’s Brachystegia Mozambique Expedition Policy spiciformis 202 Living Plant Collections: 175 Swaziland Flora Protection 212 African Botanic Gardens Mozambique Bill Congress for 2002 204 Living Plant Collections: 176 Lesotho’s State of 214 Index Herbariorum Update Namibia Environment Report 206 Living Plant Collections: 178 Marine Fishes: Are IUCN Lowveld, South Africa Red List Criteria Adequate? Book Reviews 179 Evaluating Data Deficient Taxa Against IUCN 223 Flowering Plants of the Criterion B Kalahari Dunes 180 Charcoal Production in 224 Water Plants of Namibia Malawi 225 Trees and Shrubs of the 183 Threatened -

Ecological Report on Magombera Forest
Ecological Report on Magombera Forest Andrew R. Marshall (COMMISSIONED BY WORLD WIDE FUND FOR NATURE TANZANIA PROGRAMME OFFICE) Feb 2008 2 Contents Abbreviations and Acronyms 3 Acknowledgements 4 Executive Summary 5 Background 5 Aim and Objectives 5 Findings 6 Recommendations 7 Introduction 9 Tropical Forests 9 Magombera Location and Habitat 9 Previous Ecological Surveys 10 Management and Conservation History 11 Importance of Monitoring 14 Aim and Objectives 15 Methods 15 Threats 17 Forest Structure 17 Key Species 18 Forest Restoration 20 Results and Discussion 21 Threats 21 Forest Structure 25 Key Species 26 Forest Restoration 36 Recommendations 37 Immediate Priorities 38 Short-Term Priorities 40 Long-Term Priorities 41 References 44 Appendices 49 Appendix 1. Ministry letter of support for the increased conservation of Magombera forest 49 Appendix 2. Datasheets 50 Appendix 3. List of large trees in Magombera Forest plots 55 Appendix 4. Slides used to present ecological findings to villages 58 Appendix 5. Photographs from village workshops 64 3 Abbreviations and Acronyms CEPF Critical Ecosystem Partnership Fund CITES Convention on the International Trade in Endangered Species IUCN International Union for the Conservation of Nature and Natural Resources TAZARA Tanzania-Zambia Railroad UFP Udzungwa Forest Project UMNP Udzungwa Mountains National Park WWF-TPO Worldwide Fund for Nature – Tanzania Programme Office 4 Acknowledgements Thanks to all of the following individuals and institutions: - CEPF for 2007 funds for fieldwork and report -

Following D U B a R D, the Considered Shape of the Embryo Is a Good Taxonomic Character
Notes on Guiana Sapotaceae by P. J. Eyma (Utrecht). the work Notwithstanding large amount of spent by several botanists this does not satis- on family, taxonomy appear very and has factory, a general agreement on generic limits not yet been reached. The result has been a perplexing number of generic and sectional The author for his names. present apologizes adding the number of to interpretations. This study of American Sapotaceae, primarily undertaken in connection with the Flora of Surinam, could not have been com- without the loan of the pleted generous specimens by herbaria at Brussels [B], Berlin —Dahlem [D], Kew [K], and Leyden [L]. the author short visit to the herbaria In 1934 paid a at Brussels [B] The collections and at Paris [P]. of this family at Paris are of special interest owing to the fact that they contain the material B i 11 Pierre and and studied by a on, Du'bard, bear numer- and ous notes analytical drawings, especially by Pierre, attached British to the sheets. A number of Guiana Sapotaceae from the Kew Herbarium was received for determination shortly afterwards. The author feels greatly indebted to the directors of the above their kind and mentioned Herbaria for hel{), particularly to Prof. Dr. P 11 Utrecht, under whose direction this A. u e, study was undertaken. Unless otherwise mentioned the specimens cited are in the Utrecht Herbarium [U]. The principial alterations in the classification of Sapotaceae in this due the for the paper are to rejection classifying purposes of certain number of flower-parts and, to a degree, of the staminodial development also. -

Mt Mabu, Mozambique: Biodiversity and Conservation
Darwin Initiative Award 15/036: Monitoring and Managing Biodiversity Loss in South-East Africa's Montane Ecosystems MT MABU, MOZAMBIQUE: BIODIVERSITY AND CONSERVATION November 2012 Jonathan Timberlake, Julian Bayliss, Françoise Dowsett-Lemaire, Colin Congdon, Bill Branch, Steve Collins, Michael Curran, Robert J. Dowsett, Lincoln Fishpool, Jorge Francisco, Tim Harris, Mirjam Kopp & Camila de Sousa ABRI african butterfly research in Forestry Research Institute of Malawi Biodiversity of Mt Mabu, Mozambique, page 2 Front cover: Main camp in lower forest area on Mt Mabu (JB). Frontispiece: View over Mabu forest to north (TT, top); Hermenegildo Matimele plant collecting (TT, middle L); view of Mt Mabu from abandoned tea estate (JT, middle R); butterflies (Lachnoptera ayresii) mating (JB, bottom L); Atheris mabuensis (JB, bottom R). Photo credits: JB – Julian Bayliss CS ‒ Camila de Sousa JT – Jonathan Timberlake TT – Tom Timberlake TH – Tim Harris Suggested citation: Timberlake, J.R., Bayliss, J., Dowsett-Lemaire, F., Congdon, C., Branch, W.R., Collins, S., Curran, M., Dowsett, R.J., Fishpool, L., Francisco, J., Harris, T., Kopp, M. & de Sousa, C. (2012). Mt Mabu, Mozambique: Biodiversity and Conservation. Report produced under the Darwin Initiative Award 15/036. Royal Botanic Gardens, Kew, London. 94 pp. Biodiversity of Mt Mabu, Mozambique, page 3 LIST OF CONTENTS List of Contents .......................................................................................................................... 3 List of Tables ............................................................................................................................. -

Western Lowland Gorilla (Gorilla Gorilla Gorilla) Diet and Activity Budgets: Effects of Group Size, Age Class and Food Availability in the Dzanga-Ndoki
Western lowland gorilla (Gorilla gorilla gorilla) diet and activity budgets: effects of group size, age class and food availability in the Dzanga-Ndoki National Park, Central African Republic Terence Fuh MSc Primate Conservation Dissertation 2013 Dissertation Course Name: P20107 Title: Western lowland gorilla (Gorilla gorilla gorilla) diet and activity budgets: effects of group size, age class and food availability in the Dzanga-Ndoki National Park, Central African Republic Student Number: 12085718 Surname: Fuh Neba Other Names: Terence Course for which acceptable: MSc Primate Conservation Date of Submission: 13th September 2013 This dissertation is submitted in part fulfilment of the regulations for an MSc degree. Oxford Brookes University ii Statement of originality Except for those parts in which it is explicitly stated to the contrary, this project is my own work. It has not been submitted for any degree at this or any other academic or professional institution. ……………………………………………. ………………… Signature Date Regulations Governing the Deposit and Use of Oxford Brookes University Projects/ Dissertations 1. The “top” copies of projects/dissertations submitted in fulfilment of programme requirements shall normally be kept by the School. 2. The author shall sign a declaration agreeing that the project/ dissertation be available for reading and photocopying at the discretion of the Head of School in accordance with 3 and 4 below. 3. The Head of School shall safeguard the interests of the author by requiring persons who consult the project/dissertation to sign a declaration acknowledging the author’s copyright. 4. Permission for any one other then the author to reproduce or photocopy any part of the dissertation must be obtained from the Head of School who will give his/her permission for such reproduction on to an extent which he/she considers to be fair and reasonable. -

Supplementary Material Saving Rainforests in the South Pacific
Australian Journal of Botany 65, 609–624 © CSIRO 2017 http://dx.doi.org/10.1071/BT17096_AC Supplementary material Saving rainforests in the South Pacific: challenges in ex situ conservation Karen D. SommervilleA,H, Bronwyn ClarkeB, Gunnar KeppelC,D, Craig McGillE, Zoe-Joy NewbyA, Sarah V. WyseF, Shelley A. JamesG and Catherine A. OffordA AThe Australian PlantBank, The Royal Botanic Gardens and Domain Trust, Mount Annan, NSW 2567, Australia. BThe Australian Tree Seed Centre, CSIRO, Canberra, ACT 2601, Australia. CSchool of Natural and Built Environments, University of South Australia, Adelaide, SA 5001, Australia DBiodiversity, Macroecology and Conservation Biogeography Group, Faculty of Forest Sciences, University of Göttingen, Büsgenweg 1, 37077 Göttingen, Germany. EInstitute of Agriculture and Environment, Massey University, Private Bag 11 222 Palmerston North 4474, New Zealand. FRoyal Botanic Gardens, Kew, Wakehurst Place, RH17 6TN, United Kingdom. GNational Herbarium of New South Wales, The Royal Botanic Gardens and Domain Trust, Sydney, NSW 2000, Australia. HCorresponding author. Email: [email protected] Table S1 (below) comprises a list of seed producing genera occurring in rainforest in Australia and various island groups in the South Pacific, along with any available information on the seed storage behaviour of species in those genera. Note that the list of genera is not exhaustive and the absence of a genus from a particular island group simply means that no reference was found to its occurrence in rainforest habitat in the references used (i.e. the genus may still be present in rainforest or may occur in that locality in other habitats). As the definition of rainforest can vary considerably among localities, for the purpose of this paper we considered rainforests to be terrestrial forest communities, composed largely of evergreen species, with a tree canopy that is closed for either the entire year or during the wet season. -

In Vitro Propagation of Miracle Berry (Synsepalum Dulcificum Daniel) Through Embryo and Nodal Cultures
African Journal of Biotechnology Vol. 7 (3), pp. 244-248, 5 February, 2008 Available online at http://www.academicjournals.org/AJB ISSN 1684–5315 © 2008 Academic Journals Full Length Research Paper In vitro propagation of miracle berry (Synsepalum dulcificum Daniel) through embryo and nodal cultures K. E. Ogunsola1* and C. O. Ilori2 1General Diagnostics/Biotechnology Unit, Nigeria Plant Quarantine Service PMB. 5672 Moor Plantation, Ibadan, Nigeria. 2Department of Crop Protection and Environmental Biology, University of Ibadan, Ibadan, Nigeria. Accepted 7 December, 2007 Miracle berry is an evergreen tropical shrub which modifies sour food to produce a sweet taste. Its propagation is, however, hindered by seed recalcitrance and difficulty of stem to root. Thus in vitro propagation was investigated through embryo and nodal explants using different levels and combinations of auxins and cytokinins in MS medium. Embryo was regenerated in MS medium supplemented with 0.1 mg/l NAA + 0.2 mg/l BAP. Lateral buds proliferation was induced on the germinated embryo with 0.6 - 3.0 mg/l BAP + 0.1 - 0.2 mg/l NAA in which 3.0 mg/l BAP + 0.1 mg/l NAA produced highest number of buds. Rooting of the embryo regenerated plantlets was achieved with 1.0 - 2.0 mg/l IBA + 0.1 mg/l BAP. Very low (5 - 10%) axillary and terminal buds formation was achieved from nodal cultures. Few of the nodal explants formed buds with 0.1 - 0.8 mg/l NAA + 0.2 - 1.0 mg/l BAP + 0.02 mg/l GA3 with 0.8 mg/l NAA + 0.2 mg/l BAP producing the best result. -

Perennial Edible Fruits of the Tropics: an and Taxonomists Throughout the World Who Have Left Inventory
United States Department of Agriculture Perennial Edible Fruits Agricultural Research Service of the Tropics Agriculture Handbook No. 642 An Inventory t Abstract Acknowledgments Martin, Franklin W., Carl W. Cannpbell, Ruth M. Puberté. We owe first thanks to the botanists, horticulturists 1987 Perennial Edible Fruits of the Tropics: An and taxonomists throughout the world who have left Inventory. U.S. Department of Agriculture, written records of the fruits they encountered. Agriculture Handbook No. 642, 252 p., illus. Second, we thank Richard A. Hamilton, who read and The edible fruits of the Tropics are nnany in number, criticized the major part of the manuscript. His help varied in form, and irregular in distribution. They can be was invaluable. categorized as major or minor. Only about 300 Tropical fruits can be considered great. These are outstanding We also thank the many individuals who read, criti- in one or more of the following: Size, beauty, flavor, and cized, or contributed to various parts of the book. In nutritional value. In contrast are the more than 3,000 alphabetical order, they are Susan Abraham (Indian fruits that can be considered minor, limited severely by fruits), Herbert Barrett (citrus fruits), Jose Calzada one or more defects, such as very small size, poor taste Benza (fruits of Peru), Clarkson (South African fruits), or appeal, limited adaptability, or limited distribution. William 0. Cooper (citrus fruits), Derek Cormack The major fruits are not all well known. Some excellent (arrangements for review in Africa), Milton de Albu- fruits which rival the commercialized greatest are still querque (Brazilian fruits), Enriquito D.