Isolation and Characterization of New Polymorphic Microsatellite Loci in the Mixotrophic Orchid Limodorum Abortivum L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE
Guide to the Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- LILIACEAE LILIACEAE de Jussieu 1789 (Lily Family) (also see AGAVACEAE, ALLIACEAE, ALSTROEMERIACEAE, AMARYLLIDACEAE, ASPARAGACEAE, COLCHICACEAE, HEMEROCALLIDACEAE, HOSTACEAE, HYACINTHACEAE, HYPOXIDACEAE, MELANTHIACEAE, NARTHECIACEAE, RUSCACEAE, SMILACACEAE, THEMIDACEAE, TOFIELDIACEAE) As here interpreted narrowly, the Liliaceae constitutes about 11 genera and 550 species, of the Northern Hemisphere. There has been much recent investigation and re-interpretation of evidence regarding the upper-level taxonomy of the Liliales, with strong suggestions that the broad Liliaceae recognized by Cronquist (1981) is artificial and polyphyletic. Cronquist (1993) himself concurs, at least to a degree: "we still await a comprehensive reorganization of the lilies into several families more comparable to other recognized families of angiosperms." Dahlgren & Clifford (1982) and Dahlgren, Clifford, & Yeo (1985) synthesized an early phase in the modern revolution of monocot taxonomy. Since then, additional research, especially molecular (Duvall et al. 1993, Chase et al. 1993, Bogler & Simpson 1995, and many others), has strongly validated the general lines (and many details) of Dahlgren's arrangement. The most recent synthesis (Kubitzki 1998a) is followed as the basis for familial and generic taxonomy of the lilies and their relatives (see summary below). References: Angiosperm Phylogeny Group (1998, 2003); Tamura in Kubitzki (1998a). Our “liliaceous” genera (members of orders placed in the Lilianae) are therefore divided as shown below, largely following Kubitzki (1998a) and some more recent molecular analyses. ALISMATALES TOFIELDIACEAE: Pleea, Tofieldia. LILIALES ALSTROEMERIACEAE: Alstroemeria COLCHICACEAE: Colchicum, Uvularia. LILIACEAE: Clintonia, Erythronium, Lilium, Medeola, Prosartes, Streptopus, Tricyrtis, Tulipa. MELANTHIACEAE: Amianthium, Anticlea, Chamaelirium, Helonias, Melanthium, Schoenocaulon, Stenanthium, Veratrum, Toxicoscordion, Trillium, Xerophyllum, Zigadenus. -

Phylogenetic Placement of the Enigmatic Orchid Genera Thaia and Tangtsinia: Evidence from Molecular and Morphological Characters
TAXON 61 (1) • February 2012: 45–54 Xiang & al. • Phylogenetic placement of Thaia and Tangtsinia Phylogenetic placement of the enigmatic orchid genera Thaia and Tangtsinia: Evidence from molecular and morphological characters Xiao-Guo Xiang,1 De-Zhu Li,2 Wei-Tao Jin,1 Hai-Lang Zhou,1 Jian-Wu Li3 & Xiao-Hua Jin1 1 Herbarium & State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, P.R. China 2 Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650204, P.R. China 3 Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun Township, Mengla County, Yunnan province 666303, P.R. China Author for correspondence: Xiao-Hua Jin, [email protected] Abstract The phylogenetic position of two enigmatic Asian orchid genera, Thaia and Tangtsinia, were inferred from molecular data and morphological evidence. An analysis of combined plastid data (rbcL + matK + psaB) using Bayesian and parsimony methods revealed that Thaia is a sister group to the higher epidendroids, and tribe Neottieae is polyphyletic unless Thaia is removed. Morphological evidence, such as plicate leaves and corms, the structure of the gynostemium and the micromorphol- ogy of pollinia, also indicates that Thaia should be excluded from Neottieae. Thaieae, a new tribe, is therefore tentatively established. Using Bayesian and parsimony methods, analyses of combined plastid and nuclear datasets (rbcL, matK, psaB, trnL-F, ITS, Xdh) confirmed that the monotypic genus Tangtsinia was nested within and is synonymous with the genus Cepha- lanthera, in which an apical stigma has evolved independently at least twice. -

Phylogeny, Character Evolution and the Systematics of Psilochilus (Triphoreae)
THE PRIMITIVE EPIDENDROIDEAE (ORCHIDACEAE): PHYLOGENY, CHARACTER EVOLUTION AND THE SYSTEMATICS OF PSILOCHILUS (TRIPHOREAE) A Dissertation Presented in Partial Fulfillment of the Requirements for The Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Erik Paul Rothacker, M.Sc. ***** The Ohio State University 2007 Doctoral Dissertation Committee: Approved by Dr. John V. Freudenstein, Adviser Dr. John Wenzel ________________________________ Dr. Andrea Wolfe Adviser Evolution, Ecology and Organismal Biology Graduate Program COPYRIGHT ERIK PAUL ROTHACKER 2007 ABSTRACT Considering the significance of the basal Epidendroideae in understanding patterns of morphological evolution within the subfamily, it is surprising that no fully resolved hypothesis of historical relationships has been presented for these orchids. This is the first study to improve both taxon and character sampling. The phylogenetic study of the basal Epidendroideae consisted of two components, molecular and morphological. A molecular phylogeny using three loci representing each of the plant genomes including gap characters is presented for the basal Epidendroideae. Here we find Neottieae sister to Palmorchis at the base of the Epidendroideae, followed by Triphoreae. Tropidieae and Sobralieae form a clade, however the relationship between these, Nervilieae and the advanced Epidendroids has not been resolved. A morphological matrix of 40 taxa and 30 characters was constructed and a phylogenetic analysis was performed. The results support many of the traditional views of tribal composition, but do not fully resolve relationships among many of the tribes. A robust hypothesis of relationships is presented based on the results of a total evidence analysis using three molecular loci, gap characters and morphology. Palmorchis is placed at the base of the tree, sister to Neottieae, followed successively by Triphoreae sister to Epipogium, then Sobralieae. -

Flora Mediterranea 26
FLORA MEDITERRANEA 26 Published under the auspices of OPTIMA by the Herbarium Mediterraneum Panormitanum Palermo – 2016 FLORA MEDITERRANEA Edited on behalf of the International Foundation pro Herbario Mediterraneo by Francesco M. Raimondo, Werner Greuter & Gianniantonio Domina Editorial board G. Domina (Palermo), F. Garbari (Pisa), W. Greuter (Berlin), S. L. Jury (Reading), G. Kamari (Patras), P. Mazzola (Palermo), S. Pignatti (Roma), F. M. Raimondo (Palermo), C. Salmeri (Palermo), B. Valdés (Sevilla), G. Venturella (Palermo). Advisory Committee P. V. Arrigoni (Firenze) P. Küpfer (Neuchatel) H. M. Burdet (Genève) J. Mathez (Montpellier) A. Carapezza (Palermo) G. Moggi (Firenze) C. D. K. Cook (Zurich) E. Nardi (Firenze) R. Courtecuisse (Lille) P. L. Nimis (Trieste) V. Demoulin (Liège) D. Phitos (Patras) F. Ehrendorfer (Wien) L. Poldini (Trieste) M. Erben (Munchen) R. M. Ros Espín (Murcia) G. Giaccone (Catania) A. Strid (Copenhagen) V. H. Heywood (Reading) B. Zimmer (Berlin) Editorial Office Editorial assistance: A. M. Mannino Editorial secretariat: V. Spadaro & P. Campisi Layout & Tecnical editing: E. Di Gristina & F. La Sorte Design: V. Magro & L. C. Raimondo Redazione di "Flora Mediterranea" Herbarium Mediterraneum Panormitanum, Università di Palermo Via Lincoln, 2 I-90133 Palermo, Italy [email protected] Printed by Luxograph s.r.l., Piazza Bartolomeo da Messina, 2/E - Palermo Registration at Tribunale di Palermo, no. 27 of 12 July 1991 ISSN: 1120-4052 printed, 2240-4538 online DOI: 10.7320/FlMedit26.001 Copyright © by International Foundation pro Herbario Mediterraneo, Palermo Contents V. Hugonnot & L. Chavoutier: A modern record of one of the rarest European mosses, Ptychomitrium incurvum (Ptychomitriaceae), in Eastern Pyrenees, France . 5 P. Chène, M. -

Southern Maidenhair Fern and Stream Orchid in the Black Hills National Forest, South Dakota and Wyoming
United States Department of Agriculture Conservation Assessment Forest Service for Southern Maidenhair Rocky Mountain Region Fern and Stream Orchid in Black Hills National Forest the Black Hills National Custer, South Dakota Forest South Dakota and April 2003 Wyoming J.Hope Hornbeck, Deanna Reyher, Carolyn Hull Sieg and Reed W. Crook Species Assessment of Southern Maidenhair Fern and Stream Orchid in the Black Hills National Forest, South Dakota and Wyoming J. Hope Hornbeck, Deanna J. Reyher, Carolyn Hull Sieg and Reed W. Crook J. Hope Hornbeck is a Botanist with the Black Hills National Forest in Custer, South Dakota. She completed a B.S. in Environmental Biology at The University of Montana and a M.S. in Plant Biology at the University of Minnesota-Twin Cities. Deanna J. Reyher is an Ecologist/Soil Scientist with the Black Hills National Forest in Custer, South Dakota. She completed a B.S. degree in Agronomy from the University of Nebraska- Lincoln. Carolyn Hull Sieg is a Research Plant Ecologist with the Rocky Mountain Research Station in Flagstaff, Arizona. She completed a B.S. in Wildlife Biology and M.S. in Range Science from Colorado State University and a Ph.D. in Range and Wildlife Management at Texas Tech University. Reed W. Crook is a Botanist with the Black Hills National Forest in Custer, South Dakota. He completed a B.S. in Botany at Brigham Young University, a M.S. in Plant Morphology and Ph.D. in Plant Systematics at the University of Georgia-Athens. EXECUTIVE SUMMARY Southern maidenhair fern (Adiantum capillus-veneris L.; Pteridaceae) is a cosmopolitan species that is widely distributed in southern North America. -

===Обзорные Статьи ======Orchids of Russia: Annotated Checklist and Geographic Distribution
Nature Conservation Research. Заповедная наука 2020. 5(Suppl.1): 1–18 https://dx.doi.org/10.24189/ncr.2020.018 ============== REVIEW ARTICLES ================ ============= ОБЗОРНЫЕ СТАТЬИ =============== ORCHIDS OF RUSSIA: ANNOTATED CHECKLIST AND GEOGRAPHIC DISTRIBUTION Petr G. Efimov Komarov Botanical Institute of RAS, Russia e-mail: [email protected] Received: 07.03.2020. Revised: 11.04.2020. Accepted: 13.04.2020. A checklist of orchids is presented for Russia. Data from the project «Biodiversity Mapping of Orchidaceae of Russia» are supplemented by the literature data in order to analyse the geographical distribution of species. We consider that the orchid diversity of Russia comprises 135 species and 13 subspecies belonging to 38 genera, in- cluding several taxa with status «uncertain for Russia»: Dactylorhiza majalis, D. sambucina, Epipactis purpurata, Cephalanthera caucasica, Anacamptis laxiflora subsp. palustris, and Platanthera komarovii subsp. maximovic- ziana. Krasnodarsky Krai has the highest number of orchid species (56 species), followed by Sakhalin Region (50), Republic of Dagestan (49), Republic of Crimea (45), Primorsky Krai (42), and Khabarovsky Krai (41). Only in As- trakhan Region there are no orchids known. One species (Anacamptis laxiflora, from one locality) is known from the Republic of Kalmykia, three species from Chukotsky Autonomous Okrug, eight species from Nenetsky Au- tonomous Okrug. Gymnadenia conopsea is the most widely distributed species, being found in 74 regions (90%), followed by Corallorhiza trifida (73 regions), Dactylorhiza incarnata (72), and D. viridis (71). Twenty-five species are known from only one administrative region. Eleven of them have been found in Sakhalin Region, four of them in Republic of Dagestan. Cephalanthera erecta, Epipactis euxina, and E. -

Systematics, Phylogeography, Fungal Associations, and Photosynthesis
Systematics, Phylogeography, Fungal Associations, and Photosynthesis Gene Evolution in the Fully Mycoheterotrophic Corallorhiza striata Species Complex (Orchidaceae: Epidendroideae) Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Craig Francis Barrett, M. S. Evolution, Ecology, and Organismal Biology ***** The Ohio State University 2010 Dissertation Committee: Dr. John V. Freudenstein, Advisor Dr. John W. Wenzel Dr. Andrea D. Wolfe Copyright by Craig Francis Barrett 2010 ABSTRACT Corallorhiza is a genus of obligately mycoheterotrophic (fungus-eating) orchids that presents a unique opportunity to study phylogeography, taxonomy, fungal host specificity, and photosynthesis gene evolution. The photosysnthesis gene rbcL was sequenced for nearly all members of the genus Corallorhiza; evidence for pseudogene formation was found in both the C. striata and C. maculata complexes, suggesting multiple independent transitions to complete heterotrophy. Corallorhiza may serve as an exemplary system in which to study the plastid genomic consequences of full mycoheterotrophy due to relaxed selection on photosynthetic apparatus. Corallorhiza striata is a highly variable species complex distributed from Mexico to Canada. In an investigation of molecular and morphological variation, four plastid DNA clades were identified, displaying statistically significant differences in floral morphology. The biogeography of C. striata is more complex than previously hypothesized, with two main plastid lineages present in both Mexico and northern North America. These findings add to a growing body of phylogeographic data on organisms sharing this common distribution. To investigate fungal host specificity in the C. striata complex, I sequenced plastid DNA for orchids and nuclear DNA for fungi (n=107 individuals), and found that ii the four plastid clades associate with divergent sets of ectomycorrhizal fungi; all within a single, variable species, Tomentella fuscocinerea. -

Chapter 8 DEMOGRAPHIC STUDIES and LIFE-HISTORY STRATEGIES
K.w. Dixon, S.P. Kell, R.L. Barrett and P.J. Cribb (eds) 2003. Orchid Conservation. pp. 137-158. © Natural History Publications (Borneo), Kota Kinabalu, Sabah. Chapter 8 DEMOGRAPHIC STUDIES AND LIFE-HISTORY STRATEGIES OF TEMPERATE TERRESTRIAL ORCIDDS AS A BASIS FOR CONSERVATION Dennis F Whigham Smithsonian Environmental Research Center, Box. 28, Edgewater, MD 21037, USA. Jo H. Willems Plant Ecology Group, Utrecht University, PO Box 800.84, NL 3508 Utrecht, The Netherlands. "Our knowledge about the lives ofindividual plants and oftheir persistence in plant communities is very incomplete, however, especially when perennial herbs are concerned" - C. 0. Tamm, 1948. Terrestrial orchids represent a wide diversity ofspecies that are characterised by an equally diverse range of life history attributes. Threatened and endangered species of terrestrial orchids have been identifed on all continents where they occur and conservation plans have been developed for some species. Even though there is a considerable amount of information on the ecology of terrestrial orchids, few species have been studied in detail and most management plans focus on habitat conservation. In this paper, we consider the diversity of terrestrial orchids and summarise information on threatened and endangered species from a global perspective. We also describe approaches to the conservation and restoration ofterrestrial orchids and develop the argument that much information is needed ifwe are to successfully conserve this diverse group ofplant species. 1. Introduction Terrestrial orchids represent a wide variety of life history types, from autotrophic evergreen to completely myco-heterotrophic species that obtain most oftheir resources from a mycobiont. Life history characteristics ofterrestrial orchids are generally well known (e.g. -
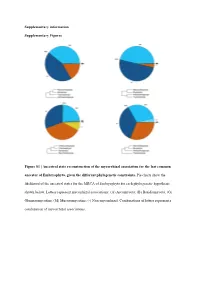
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Nutritional Regulation in Mixotrophic Plants: New Insights from Limodorum Abortivum
Oecologia (2014) 175:875–885 DOI 10.1007/s00442-014-2940-8 PLANT-MICROBE-ANIMAL INTERACTIONS - ORIGINAL RESEARCH Nutritional regulation in mixotrophic plants: new insights from Limodorum abortivum Alessandro Bellino · Anna Alfani · Marc-André Selosse · Rossella Guerrieri · Marco Borghetti · Daniela Baldantoni Received: 15 January 2014 / Accepted: 28 March 2014 / Published online: 11 May 2014 © Springer-Verlag Berlin Heidelberg 2014 Abstract Partially mycoheterotrophic (mixotrophic) about this nutrition, and especially about the physiologi- plants gain carbon from both photosynthesis and their cal balance between photosynthesis and fungal C gain. To mycorrhizal fungi. This is considered an ancestral state in investigate possible compensation between photosynthesis the evolution of full mycoheterotrophy, but little is known and mycoheterotrophy in the Mediterranean mixotrophic orchid Limodorum abortivum, fungal colonization was experimentally reduced in situ by fungicide treatment. We Electronic supplementary material The online version of this article (doi:10.1007/s00442-014-2940-8) contains supplementary measured photosynthetic pigments of leaves, stems, and material, which is available to authorized users. ovaries, as well as the stable C isotope compositions (a proxy for photosynthetic C gain) of seeds and the sizes of Communicated by Caroline Müller. ovaries and seeds. We demonstrate that (1) in natural con- Alessandro Bellino began this research, then Daniela Baldantoni ditions, photosynthetic pigments are most concentrated worked alongside him in every phase of this project, from in ovaries; (2) pigments and photosynthetic C increase devising the experimental plan to the samplings, analyses, and in ovaries when fungal C supply is impaired, buffering C interpretation of data. Marc-André Selosse proposed additional limitations and allowing the same development of ova- experiments and measurements, and carried out some of them in his lab. -

The Molecular and Morphological Systematics of Subfamily Epidendroideae (Orchidaceae)
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1995 The olecM ular and Morphological Systematics of Subfamily Epidendroideae (Orchidaceae). Malcolm Ray Neyland Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Neyland, Malcolm Ray, "The oM lecular and Morphological Systematics of Subfamily Epidendroideae (Orchidaceae)." (1995). LSU Historical Dissertations and Theses. 6040. https://digitalcommons.lsu.edu/gradschool_disstheses/6040 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely afreet reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. -

Comparative Vascular Anatomies of Some Orchid Species Bazı Orkide
Süngü Şeker et al – Comparative vascular anatomies … Anatolian Journal of Botany Anatolian Journal of Botany 5(2): 84-90 (2021) Research article doi:10.30616/ajb.905956 Comparative vascular anatomies of some orchid species Şenay SÜNGÜ ŞEKER1 , Gülcan ŞENEL2* , Mustafa Kemal AKBULUT3 1,2Ondokuz Mayıs University, Sciences and Arts Faculty, Department of Biology, Samsun, Turkey 3 Çanakkale Onsekiz Mart University, Lapseki Vocational School, Department of Landscaping and Ornamental Plants, Çanakkale, Turkey *[email protected], [email protected], [email protected] Received : 30.03.2021 Accepted : 10.06.2021 Bazı orkide türlerinin karşılaştırmalı damar anatomileri Online : 22.06.2021 Abstract: In this study, we examined the vascular anatomy of leaves with different morphological features of 11 orchid species. Plant samples were collected from various localities in the Black Sea Region. Fresh leaves were dried and stocked, and their vascular structures were analyzed by clearing and staining. Significant differences were determined in the leaves of taxa in terms of characters such as total leaf perimeter and area, number of veins and nodes, total vein length, total vein area, average vein length, average vein width, average vein surface area, average vein volume, and average areolar area. According to the findings, the topological and morphometric features of the veining can reflect the systematic and phylogenetic relationships of orchids. Key words: Anatomy, venation network, morphometry, Orchidaceae Özet: Bu çalışmada 11 orkide türüne ait farklı morfolojik özellikleri olan yaprakların damar anatomileri incelenmiştir. Bitki örnekleri Karadeniz Bölgesi’ndeki çeşitli lokalitelerden toplanmıştır. Taze yapraklar kurutularak stoklanmış, saydamlaştırma ve boyama işlemi uygulanarak damar yapıları analiz edilmiştir.