Phytosociology and Genetic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Universite Toulouse Iii Paul Sabatier Faculte Des Sciences Pharmaceutiques
UNIVERSITE TOULOUSE III PAUL SABATIER FACULTE DES SCIENCES PHARMACEUTIQUES ANNEE: 2015 THESES 2015 TOU3 2118 THESE POUR LE DIPLOME D'ETAT DE DOCTEUR EN PHARMACIE Présentée et soutenue publiquement par HAMEG LISA LES JARDINS BOTANIQUES DANS LES FACULTES DE PHARMACIE FRANCAISES - PARTICIPATION AU PROJET SMARTJARDIN Vendredi 18 décembre 2015 Directeur de thèse : Vansteelandt Marieke JURY Président : Fabre, Nicolas 1er assesseur : Vansteelandt, Marieke 2ème assesseur : Chosson, Elizabeth PERSONNEL ENSEIGNANT de la Faculté des Sciences Pharmaceutiques de l’Université Paul Sabatier au 1er octobre 2015 Professeurs Émérites M. BASTIDE R Pharmacie Clinique M. BERNADOU J Chimie Thérapeutique M. CAMPISTRON G Physiologie M. CHAVANT L Mycologie Mme FOURASTÉ I Pharmacognosie M. MOULIS C Pharmacognosie M. ROUGE P Biologie Cellulaire Professeurs des Universités Hospitalo-Universitaires Universitaires M. CHATELUT E Pharmacologie Mme BARRE A Biologie M. FAVRE G Biochimie Mme BAZIARD G Chimie pharmaceutique M. HOUIN G Pharmacologie Mme BENDERBOUS S Mathématiques – Biostat. M. PARINI A Physiologie M. BENOIST H Immunologie M. PASQUIER C (Doyen) Bactériologie - Virologie Mme BERNARDES-GÉNISSON V Chimie thérapeutique Mme ROQUES C Bactériologie - Virologie Mme COUDERC B Biochimie Mme ROUSSIN A Pharmacologie M. CUSSAC D (Vice-Doyen) Physiologie Mme SALLERIN B Pharmacie Clinique Mme DOISNEAU-SIXOU S Biochimie M. SIÉ P Hématologie M. FABRE N Pharmacognosie M. VALENTIN A Parasitologie M. GAIRIN J-E Pharmacologie Mme MULLER-STAUMONT C Toxicologie - Sémiologie Mme NEPVEU F Chimie analytique M. SALLES B Toxicologie M. SÉGUI B Biologie Cellulaire M. SOUCHARD J-P Chimie analytique Mme TABOULET F Droit Pharmaceutique M. VERHAEGHE P Chimie Thérapeutique Maîtres de Conférences des Universités Hospitalo-Universitaires Universitaires M. CESTAC P Pharmacie Clinique Mme ARÉLLANO C. -

Taxonomic Revision of Flora of Rawalakot, District Poonch Azad Jammu and Kashmir
TAXONOMIC REVISION OF FLORA OF RAWALAKOT, DISTRICT POONCH AZAD JAMMU AND KASHMIR Sajjad Hussain (Regd. No. 99-GRTB-1475) Session: 2009-12 Department of Botany Faculty of Science and Technology University of Azad Jammu and Kashmir, Muzaffarabad 1 2 TAXONOMIC REVISION OF FLORA OF RAWALAKOT, DISTRICT POONCH AZAD JAMMU AND KASHMIR By Sajjad Hussain (Regd. No. 99-GRTB-1475) A dissertation Submitted in partial fulfillment of the requirements for the degree of Doctor of philosophy In Botany Session 2009-2012 Department of Botany Faculty of Sciences and Technology University of Azad Jammu and Kashmir, Muzaffarabad 3 4 Dedicated to MY MOTHER WHO HAS ALWAYS BEEN AN INSPIRATION FOR ME 5 CONTENTS Page Acknowledgements xii List of Tables ix List of Figures ix List of Plates ix Abstract xiii 1. INTRODUCTION 1 2. REVIEW OF LITERATURE 6 3. MATERIALS AND METHODS 15 4. RESULTS AND DISCUSSION 22 4.1. Results 22 4.1.1. Checklist of angiosperm of Rawalakot 22 4.1.2. Flora of Rawalakot 39 4.1.2.1. Acanthaceae 39 4.1.2.2. Aceraceae 41 4.1.2.3. Amaranthaceae 41 4.1.2.4. Apiaceae 44 4.1.2.5. Apocynaceae 46 4.1.2.6. Araliaceae 47 4.1.2.7. Aristolochiaceae 48 4.1.2.8. Asclepiadaceae 48 4.1.2.9. Asteraceae 48 4.1.2.10. Balsaminaceae 66 4.1.2.11. Berberidaceae 67 6 4.1.2.12. Boraginaceae 68 4.1.2.13. Brassicaceae 69 4.1.2.14. Buddlejaceae 75 4.1.2.15. Buxaceae 75 4.1.2.16. -

Check List Lists of Species Check List 12(1): 1824, 6 January 2016 Doi: ISSN 1809-127X © 2016 Check List and Authors
12 1 1824 the journal of biodiversity data 6 January 2016 Check List LISTS OF SPECIES Check List 12(1): 1824, 6 January 2016 doi: http://dx.doi.org/10.15560/12.1.1824 ISSN 1809-127X © 2016 Check List and Authors Flora of Niti Valley: a cold arid region of Nanda Devi Biosphere Reserve, Western Himalaya, India Amit Kumar, Monideepa Mitra, Bhupendra S. Adhikari* and Gopal S. Rawat Department of Habitat Ecology, Wildlife Institute of India, Post Box #18, Chandrabani, Dehradun 248001, Uttarakhand, India * Corresponding author. E-mail: [email protected] Abstract: Located in the extended buffer zone of region and characterized by extreme climatic conditions, Nanda Devi Biosphere Reserve in Western Himalaya, such as diurnal fluctuations in temperatures, scanty and Niti valley represents a cold arid region. The reserve has erratic rainfall, heavy winds and snowfall. been extensively surveyed in terms of floral diversity by The Indian Trans-Himalaya (ITH) usually described as various workers, albeit highly confined to the core zones. ‘High Altitude Cold Desert Zone’ (Zone 1) spreads into The current survey recorded 495 species belonging to three biogeographic provinces: 1A, Ladakh mountains: 267 genera and 73 families of vascular plants through Kargil, Nubra and Zanskar in Jammu and Kashmir systematic collection in the years 2011, 2012 and 2014. and Lahul and Spiti in Himachal Pradesh); 1B, Tibetan Of the recorded species, 383 were dicots, 93 monocots, plateau: Changthang region of Ladakh and northern 9 pteridophytes and 10 gymnosperms. Asteraceae was parts of the states of Uttarakhand; and 1C, Sikkim most diverse family (32 genera with 58 species), followed Plateau (Rodgers et al. -

Additions to the Floral Wealth of Sirmaur District, Himachal Pradesh from Churdhar Wildlife Sanctuary
Journal of Threatened Taxa | www.threatenedtaxa.org | 26 October 2014 | 6(11): 6427–6452 Additions to the floral wealth of Sirmaur District, Himachal Pradesh from Churdhar Wildlife Sanctuary 1 2 3 ISSN 0974-7907 (Online) S.P. Subramani , K.S. Kapoor & G.S. Goraya Communication Short ISSN 0974-7893 (Print) 1,2,3 Himalayan Forest Research Institute (HFRI), Conifer Campus, Panthaghati, Shimla, Himachal Pradesh 171009, India 1 Present Address: Institute of Forest Genetics and Tree Breeding (IFGTB), Forest Campus, R.S. Puram, Coimbatore, OPEN ACCESS Tamil Nadu 641002, India 3 Present Address: Deputy Director General, Indian Council of Foretry Research & Education (ICFRE), Dehra Dun, Uttarkhand 248006, India 1 [email protected] (corresponding author), 2 [email protected], 3 [email protected] Abstract: A total of 352 species of phenerogams comprising 251 use of natural resources. As a result, they store more dicotyledons, 97 monocotyledons and four species of gymnosperms than 15% of the terrestrial carbon (Holdgate 1999) and belonging to 85 families collected from Nohra Forest Block of Churdhar Wildlife Sanctuary, District Sirmaur, Himachal Pradesh are reported as also serve as a tool for containing future exceptional additions to the district. This includes 13 threatened species under loss of habitat. Accordingly, it has now been realized different Red List assessments, both global and regional and 35 species are endemic to western Himalaya. that inventorisation of plants occurring in such areas is absolutely necessary and so are the initiatives from Keywords: Additions, Churdhar, flora, Himachal Pradesh, protected the Ministry of Environment, Forests, & Climate Change, area, Sirmaur. Government of India, which is now showing great concern for compilation or documentation of floral elements in these areas of importance, ultimately leading towards Protected areas (PAs) like wildlife reserves and ensuring the welfare of faunal elements. -

Download Download
Firenze University Press Caryologia www.fupress.com/caryologia International Journal of Cytology, Cytosystematics and Cytogenetics Pollen and floral micromorphological Studies of the genus Cotoneaster Medik. (Rosaceae) and Citation: N. Allah Raei Niaki, F. Attar, M. Mirtadzadini, K. Mahdigholi (2020) Pol- its systematic importance len and floral micromorphological Stud- ies of the genus Cotoneaster Medik. (Rosaceae) and its systematic impor- tance. Caryologia 73(3): 133-151. doi: Nemat Allah Raei Niaki1, Farideh Attar1,*, Mansour Mirtadzadini2, 10.13128/caryologia-569 Kazem Mahdigholi1 Received: July 18, 2019 1Central Herbarium of Tehran University, School of Biology, College of Science, Univer- sity of Tehran, Iran Accepted: July 16, 2020 2 Deptartment of Biology, Faculty of Science, Shahid Bahonar University, PO Box 76169- Published: December 31, 2020 133, Kerman, Iran * Corresponding author. E-mail: [email protected]) Copyright: © 2020 N. Allah Raei Niaki, F. Attar, M. Mirtadzadini, K. Mahdigholi. This is an open access, peer-reviewed Abstract. The micromorphology of petal and pollen grain of 16 species of the genus article published by Firenze University Cotoneaster Medik., belonging to two subgenera (Cotoneaster and Chaenopetalum) has Press (http://www.fupress.com/caryo- been studied by light and scanning electron microscopy. We used different multivariate logia) and distributed under the terms statistical methods to reveal the species relationships. Results showed that in compari- of the Creative Commons Attribution son to most genera of the family Rosaceae, both tri- and tetracolporate pollen grains are License, which permits unrestricted observed in one specimen. Palynological observation revealed that shape of tricolpo- use, distribution, and reproduction in any medium, provided the original rate pollen in most species is prolate-spheroidal, but also subprolate and prolate pollen author and source are credited. -

UNIVERSITY of PÉCS Biogeographic Perspectives Of
UNIVERSITY OF PÉCS Doctoral School of Biology and Sport Biology Biogeographic perspectives of Jerusalem artichoke (Helianthus tuberosus L. s. l.) invasion PhD Thesis Rita Filep PÉCS, 2018 UNIVERSITY OF PÉCS Doctoral School of Biology and Sport Biology Biogeographic perspectives of Jerusalem artichoke (Helianthus tuberosus L. s. l.) invasion PhD Thesis Rita Filep Supervisors …………………………… …………………………… Dr. Ágnes Farkas Dr. Róbert Pál associate professor assistant professor Program Director …………………………… Dr. Róbert Gábriel full professor PÉCS, 2018 ‘It is not the strongest of the species that survives, nor the most intelligent that survives. It is the one that is most adaptable to change.’ Charles Darwin I dedicate this dissertation to my family, who have taught me to work hard to achieve my goals. TABLE OF CONTENTS List of figures ................................................................................................................1 List of tables ..................................................................................................................3 1. MOTIVATION ......................................................................................................4 2. INTRODUCTION..................................................................................................5 2.1. Plant invasion ..................................................................................................5 2.1.1. Introduction of alien plants .......................................................................5 2.1.2. The process -

Quarterly Changes
Plant Names Database: Quarterly changes 31 August 2017 © Landcare Research New Zealand Limited 2017 This copyright work is licensed under the Creative Commons Attribution 4.0 license. Attribution if redistributing to the public without adaptation: "Source: Landcare Research" Attribution if making an adaptation or derivative work: "Sourced from Landcare Research" http://dx.doi.org/doi:10.7931/P13K9N CATALOGUING IN PUBLICATION Plant names database: quarterly changes [electronic resource]. – [Lincoln, Canterbury, New Zealand] : Landcare Research Manaaki Whenua, 2014- . Online resource Quarterly November 2014- ISSN 2382-2341 I.Manaaki Whenua-Landcare Research New Zealand Ltd. II. Allan Herbarium. Citation and Authorship Wilton, A.D.; Schönberger, I.; Gibb, E.S.; Boardman, K.F.; Breitwieser, I.; Cochrane, M.; Dawson, M.I.; de Pauw, B.; Fife, A.J.; Ford, K.A.; Glenny, D.S.; Heenan, P.B.; Korver, M.A.; Novis, P.M.; Redmond, D.N.; Smissen, R.D. Tawiri, K. (2017) Plant Names Database: Quarterly changes. August 2017. Lincoln, Manaaki Whenua Press. This report is generated using an automated system and is therefore authored by the staff at the Allan Herbarium who currently contribute directly to the development and maintenance of the Plant Names Database. Authors are listed alphabetically after the third author. Authors have contributed as follows: Leadership: Wilton, Heenan, Breitwieser Database editors: Wilton, Schönberger, Gibb Taxonomic and nomenclature research and review: Schönberger, Gibb, Wilton, Breitwieser, Dawson, Ford, Fife, Glenny, Heenan, Novis, Redmond, Smissen Information System development: Wilton, De Pauw, Cochrane Technical support: Boardman, Korver, Redmond, Tawiri Disclaimer The Plant Names Database is being updated every working day. We welcome suggestions for improvements, concerns, or any data errors you may find. -
UNIVERSITY of PÉCS Biogeographic Perspectives of Jerusalem Artichoke (Helianthus Tuberosus L. S. L.) Invasion Rita Filep PÉCS
UNIVERSITY OF PÉCS Doctoral School of Biology and Sport Biology Biogeographic perspectives of Jerusalem artichoke (Helianthus tuberosus L. s. l.) invasion Theses of PhD Dissertation Rita Filep Supervisors: Dr. Ágnes Farkas Dr. Róbert Pál PhD, habil. PhD, habil. PÉCS, 2018 1. Introduction Due to direct and indirect consequences of human activities, about 6.2 alien species arrive to Europe from other continents every year (Pyšek et al. 2004; Lambdon et al. 2008). The majority of plants have been introduced as ornamentals or cultivated species (Lambdon et al. 2008). However, some exotic species escaped cultivation and became subspontaneous agricultural weeds or invaders at various native ecosystems causing serious environmental problems (Kovács 2006). The impact of plant invasion falls into broad categories: starting with the environment, through human or animal health, as far as economic. Within the environment category, ecological impacts are the most difficult to quantify (Barney et al. 2013), because they depend on the attributes of recipient ecosystems and the invaders themselves (Levine et al. 2003). In the last few decades invasive exotic plants have become the most serious actual causes of species number declines and native habitat degradation (Vilà et al 2006; Mollot et al. 2017). Thus, invasive alien plant species have been recognized as one of the potential threats to native plant diversity (Corlett 2016) through reduction of genetic variation via hybridization, facilitation of pathogen spread, parasitism, and predation (Callaway and Maron 2006). Furthermore, the abundance and ecological impacts of some invasive plant species are much greater in their non-native ranges than in their native ranges (Callaway et al. -

Ploidy Levels, Relative Genome Sizes, and Base Pair Composition in Cotoneaster
J. AMER.SOC.HORT.SCI. 141(5):457–466. 2016. doi: 10.21273/JASHS03776-16 Ploidy Levels, Relative Genome Sizes, and Base Pair Composition in Cotoneaster Joseph J. Rothleutner Lincoln Park Zoo, 2001 North Clark Street, Chicago, IL 60614 Mara W. Friddle and Ryan N. Contreras1 Department of Horticulture, Oregon State University, 4017 Agricultural and Life Sciences Building, Corvallis, OR 97331 ADDITIONAL INDEX WORDS. Rosaceae, Maloideae, chromosome number, flow cytometry ABSTRACT. The genus Cotoneaster (Rosaceae, Maloideae) is highly diverse, containing ’400 species. Like other maloids, there is a high frequency of naturally occurring polyploids within the genus, with most species being tetraploid or triploid. Apomixis is also prevalent and is associated with polyploidy. The objective of this study was to estimate genome sizes and infer ploidy levels for species that had not previously been investigated as well as compare estimates using two fluorochromes and determine base pair (bp) composition. Chromosome counts of seven species confirmed ploidy levels estimated from flow cytometric analysis of nuclei stained with 4#,6-diamidino-2-phenylindole (DAPI). Monoploid (1Cx) genome sizes ranged from 0.71 to 0.96 pg. Differences in monoploid genome size were not related to current taxonomic treatment, indicating that while chromosome sizes may vary among species, there are no clear differences related to subgeneric groups. A comparison of DAPI and propidium iodide (PI) showed a difference in DNA staining in Cotoneaster comparable to other rosaceous species. Base pair composition (AT%) in Cotoneaster ranged from 58.4% to 60.8%, which led to overestimation of genome size estimates in many cases—assuming the estimates of the DNA intercalator are accurate. -
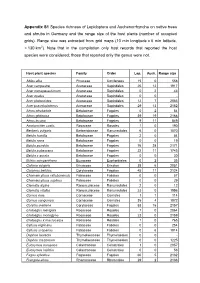
Appendix S1 Species Richness of Lepidoptera and Auchenorrhyncha on Native Trees and Shrubs in Germany and the Range Size of the Host Plants (Number of Occupied Grids)
Appendix S1 Species richness of Lepidoptera and Auchenorrhyncha on native trees and shrubs in Germany and the range size of the host plants (number of occupied grids). Range size was extracted from grid maps (10 min longitude x 6 min latitude, ≈ 130 km 2). Note that in the compilation only host records that reported the host species were considered; those that reported only the genus were not. Host plant species Family Order Lep. Auch. Range size Abies alba Pinaceae Coniferales 15 0 558 Acer campestre Aceraceae Sapindales 25 12 1917 Acer monspessulanum Aceraceae Sapindales 0 2 43 Acer opalus Aceraceae Sapindales 0 0 - Acer platanoides Aceraceae Sapindales 12 7 2083 Acer pseudoplatanus Aceraceae Sapindales 29 13 2152 Alnus alnobetula Betulaceae Fagales 0 2 54 Alnus glutinosa Betulaceae Fagales 39 19 2168 Alnus incana Betulaceae Fagales 9 11 849 Amelanchier ovalis Rosaceae Rosales 1 0 190 Berberis vulgaris Berberidaceae Ranunculales 6 0 1070 Betula humilis Betulaceae Fagales 2 0 54 Betula nana Betulaceae Fagales 0 0 19 Betula pendula Betulaceae Fagales 76 28 2171 Betula pubescens Betulaceae Fagales 22 11 1745 Betula x aurata Betulaceae Fagales 0 0 30 Buxus sempervirens Buxaceae Euphorbiales 0 2 35 Calluna vulgaris Ericaceae Ericales 38 6 2051 Carpinus betulus Corylaceae Fagales 45 11 2124 Chamaecytisus ratisbonensis Fabaceae Fabales 0 0 57 Chamaecytisus supinus Fabaceae Fabales 0 0 29 Clematis alpina Ranunculaceae Ranunculales 2 0 12 Clematis vitalba Ranunculaceae Ranunculales 23 0 1556 Cornus mas Cornaceae Cornales 1 1 114 Cornus sanguinea -

Global Flora Vol 4
THE GLOBAL FLORA © 2018 Plant Gateway Ltd. A practical flora to vascular plant species of the world ISSN 2398-6336 eISSN 2398-6344 www.plantgateway.com/globalflora/ eISBN 978-0-9929993-6-0 i Published online 9 February 2018 PLANT GATEWAy’s THE GLOBAL FLORA A practical flora to vascular plant species of the world GLOVAP Nomenclature Part 1 February 2018 The Global Flora A practical flora to vascular plant species of the world Special Edition, GLOVAP Nomenclature Part 1, Vol. 4: 1-155. Published by Plant Gateway Ltd., 5 Baddeley Gardens, Bradford, BD10 8JL, United Kingdom © Plant Gateway 2018 This work is in copyright. Subject to statutory exception and to the provision of relevant col- lective licensing agreements, no reproduction of any part may take place without the written permission of Plant Gateway Ltd. ISSN 2398-6336 eISSN 2398-6344 ISBN 978-0-9929993-5-3 eISBN 978-0-9929993-6-0 Plant Gateway has no responsibility for the persistence or accuracy of URLS for external or third-party internet websites referred to in this work, and does not guarantee that any con- tent on such websites is, or will remain, accurate or appropriate. British Library Cataloguing in Publication data A Catalogue record of this book is available from the British Library For information or to purchase other Plant Gateway titles please visit www.plantgateway.com Cover image: Hakea plurijuga (F.Muell.) Christenh. & Byng © Maarten Christenhusz Editors Maarten J.M. Christenhusz Plant Gateway, Bradford & Kingston, United Kingdom and Den Haag, the Netherlands Royal Botanic Gardens, Kew, United Kingdom. [email protected] Michael F. -

Species of Cotoneaster (Rosaceae, Maloideae) Indigenous To, Naturalising Or Common- Ly Cultivated in Central Europe
Willdenowia 40 – 2010 13 Wolf Bernhard dickoré1* & GerWin kasperek2 Species of Cotoneaster (Rosaceae, Maloideae) indigenous to, naturalising or common- ly cultivated in Central Europe Abstract dickoré W. B. & kasperek G.: species of Cotoneaster (Rosaceae, Maloideae) indigenous to, naturalising or com- monly cultivated in central europe. – Willdenowia 40: 13–45. – online issn 1868-6397; © 2010 BGBM Berlin- dahlem. doi:10.3372/wi.40.40102 (available via http://dx.doi.org/) several alien species of the eurasian genus Cotoneaster are naturalising in central europe, apparently increasingly so, and some on a massive scale. They presumably originate from large-scale cultivation for ground cover, hedges or as ornamental shrubs. The present paper keys and synopses the Cotoneaster species indigenous to, naturalis- ing or commonly cultivated in central europe, on the basis of, relatively limited, both living (wild, adventive and cultivated) and herbarium material. an attempt is made to understand the nature of variation from the genus’ centre of diversification, the mountains of china and the himalayas, which are likewise the origin of most cultivated and naturalising Cotoneaster species. Taxonomic and nomenclatural problems, putatively relating to the presence of apomixis and hybridization in the genus, are discussed. Many of the more than 500 published binomials, including a substantial proportion of those based on cultivated material, seem to be poorly defined, both morphologically and chorologically. of an estimated total of only 50–70 Cotoneaster species worldwide, about 20, mainly chinese spe- cies have been found escaping from cultivation in central europe. presently, about ten species must be considered fully naturalised and, locally at least, invasive.