1412-033X (Printed Edition) ISSN: 2085-4722 (Electronic)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Rhacophoridae, Pseudophilautus) in Sri Lanka
Molecular Phylogenetics and Evolution 132 (2019) 14–24 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Diversification of shrub frogs (Rhacophoridae, Pseudophilautus) in Sri Lanka T – Timing and geographic context ⁎ Madhava Meegaskumburaa,b,1, , Gayani Senevirathnec,1, Kelum Manamendra-Arachchid, ⁎ Rohan Pethiyagodae, James Hankenf, Christopher J. Schneiderg, a College of Forestry, Guangxi Key Lab for Forest Ecology and Conservation, Guangxi University, Nanning 530004, PR China b Department of Molecular Biology & Biotechnology, Faculty of Science, University of Peradeniya, Peradeniya, Sri Lanka c Department of Organismal Biology & Anatomy, University of Chicago, Chicago, IL, USA d Postgraduate Institute of Archaeology, Colombo 07, Sri Lanka e Ichthyology Section, Australian Museum, Sydney, NSW 2010, Australia f Museum of Comparative Zoology, Harvard University, Cambridge, MA 02138, USA g Department of Biology, Boston University, Boston, MA 02215, USA ARTICLE INFO ABSTRACT Keywords: Pseudophilautus comprises an endemic diversification predominantly associated with the wet tropical regions ofSri Ancestral-area reconstruction Lanka that provides an opportunity to examine the effects of geography and historical climate change on diversi- Biogeography fication. Using a time-calibrated multi-gene phylogeny, we analyze the tempo of diversification in thecontextof Ecological opportunity past climate and geography to identify historical drivers of current patterns of diversity and distribution. Molecular Diversification dating suggests that the diversification was seeded by migration across a land-bridge connection from India duringa Molecular dating period of climatic cooling and drying, the Oi-1 glacial maximum around the Eocene-Oligocene boundary. Lineage- Speciation through-time plots suggest a gradual and constant rate of diversification, beginning in the Oligocene and extending through the late Miocene and early Pliocene with a slight burst in the Pleistocene. -

Species Account
REPTILIA Order OPHIDIA (Snakes) I. Family COLUBRIDAE Ahaetulla prasina Green Vine Snake This snake was found in Renah Kayu Embun and Napal Licin survey sites at elevation 1400 meters asl and 300 meters asl respectively. Usually it can be seen in degraded habitat including plantation, secondary growth and house compounds, to primary rain forest (Inger and Stuebing, 2005; Kurniati, 2003). It occurs from lowlands up to mountain forests over 1500 meters asl (Kurniati et al., 2001; Kurniati, 2003). It is common species at low elevation (Inger and Stuebing, 2005), but become rare at high elevation such as Renah Kayu Embun survey site. This species is known from South-east Asia, East Indies (Sulawesi and The Lesser Sunda) (Stuebing and Inger, 1999: de Lang and Vogel, 2005). Figure 91. A. prasina (Photograph by H. Kurniati). Amphiesma sp This undescribed snake was found in Muara Labuh survey site at elevation 800 meters asl. It was a nocturnal snake that inhabited strong moving stream bank. The morphology of this snake is similar to A. kerinciense (David and Das, 2003). Possibly, it is a new species, but future study is needed. Figure 92. Amphiesma sp from Muara Labuh (Photograph by H. Kurniati). Aplopeltura boa Blunt-headed Tree Snake This snake was found in Upper Rupit River and Tapan survey sites at elevation 150 meters asl and 550 meters asl respectively. It inhabited lowland primary rain forest. It occurs at elevation between sea level to 1200 meters asl (Kurniati, 2003), but it is confined to be lowland. In Tapan survey site, it was rarely observed. -

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

Download Download
HAMADRYAD Vol. 27. No. 2. August, 2003 Date of issue: 31 August, 2003 ISSN 0972-205X CONTENTS T. -M. LEONG,L.L.GRISMER &MUMPUNI. Preliminary checklists of the herpetofauna of the Anambas and Natuna Islands (South China Sea) ..................................................165–174 T.-M. LEONG & C-F. LIM. The tadpole of Rana miopus Boulenger, 1918 from Peninsular Malaysia ...............175–178 N. D. RATHNAYAKE,N.D.HERATH,K.K.HEWAMATHES &S.JAYALATH. The thermal behaviour, diurnal activity pattern and body temperature of Varanus salvator in central Sri Lanka .........................179–184 B. TRIPATHY,B.PANDAV &R.C.PANIGRAHY. Hatching success and orientation in Lepidochelys olivacea (Eschscholtz, 1829) at Rushikulya Rookery, Orissa, India ......................................185–192 L. QUYET &T.ZIEGLER. First record of the Chinese crocodile lizard from outside of China: report on a population of Shinisaurus crocodilurus Ahl, 1930 from north-eastern Vietnam ..................193–199 O. S. G. PAUWELS,V.MAMONEKENE,P.DUMONT,W.R.BRANCH,M.BURGER &S.LAVOUÉ. Diet records for Crocodylus cataphractus (Reptilia: Crocodylidae) at Lake Divangui, Ogooué-Maritime Province, south-western Gabon......................................................200–204 A. M. BAUER. On the status of the name Oligodon taeniolatus (Jerdon, 1853) and its long-ignored senior synonym and secondary homonym, Oligodon taeniolatus (Daudin, 1803) ........................205–213 W. P. MCCORD,O.S.G.PAUWELS,R.BOUR,F.CHÉROT,J.IVERSON,P.C.H.PRITCHARD,K.THIRAKHUPT, W. KITIMASAK &T.BUNDHITWONGRUT. Chitra burmanica sensu Jaruthanin, 2002 (Testudines: Trionychidae): an unavailable name ............................................................214–216 V. GIRI,A.M.BAUER &N.CHATURVEDI. Notes on the distribution, natural history and variation of Hemidactylus giganteus Stoliczka, 1871 ................................................217–221 V. WALLACH. -

Conservation of Amphibians and Reptiles in Indonesia: Issues and Problems
Copyright: © 2006 Iskandar and Erdelen. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and repro- Amphibian and Reptile Conservation 4(1):60-87. duction in any medium, provided the original author and source are credited. DOI: 10.1514/journal.arc.0040016 (2329KB PDF) The authors are responsible for the facts presented in this article and for the opinions expressed there- in, which are not necessarily those of UNESCO and do not commit the Organisation. The authors note that important literature which could not be incorporated into the text has been published follow- ing the drafting of this article. Conservation of amphibians and reptiles in Indonesia: issues and problems DJOKO T. ISKANDAR1 * AND WALTER R. ERDELEN2 1School of Life Sciences and Technology, Institut Teknologi Bandung, 10, Jalan Ganesa, Bandung 40132 INDONESIA 2Assistant Director-General for Natural Sciences, UNESCO, 1, rue Miollis, 75732 Paris Cedex 15, FRANCE Abstract.—Indonesia is an archipelagic nation comprising some 17,000 islands of varying sizes and geologi- cal origins, as well as marked differences in composition of their floras and faunas. Indonesia is considered one of the megadiversity centers, both in terms of species numbers as well as endemism. According to the Biodiversity Action Plan for Indonesia, 16% of all amphibian and reptile species occur in Indonesia, a total of over 1,100 species. New research activities, launched in the last few years, indicate that these figures may be significantly higher than generally assumed. Indonesia is suspected to host the worldwide highest numbers of amphibian and reptiles species. -

First National Record of <I>Gracixalus Quangi</I>
Biodiversity Data Journal 9: e67667 doi: 10.3897/BDJ.9.e67667 Taxonomic Paper First national record of Gracixalus quangi Rowley, Dau, Nguyen, Cao & Nguyen, 2011 and G. yunnanensis Yu, Li, Wang, Rao, Wu &Yang, 2019 (Amphibia: Anura: Rhacophoridae) from Thailand Sengvilay Lorphengsy‡,§, Tan Van Nguyen|, Nikolay A. Poyarkov¶,#, Yun-He Wu ¤, Parinya Pawangkhanant«, Supaporn Passorn‡, Jing Che ¤, Chatmongkon Suwannapoom« ‡ Division of Biotechnology, School of Agriculture and Natural Resources, University of Phayao, Phayao, Thailand § The Biotechnology and Ecology Institute Ministry of Science and Technology, Vientiane, Laos | Department of Species Conservation, Save Vietnam’s Wildlife,, Ninh Binh, Vietnam ¶ Faculty of Biology, Department of Vertebrate Zoology, Moscow State University, Moscow, Moscow, Russia # Laboratory of Tropical Ecology, Joint Russian-Vietnamese Tropical Research and Technological Center, Hanoi, Vietnam ¤ State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China « Division of Fishery, School of Agriculture and Natural Resources, University of Phayao, Phayao, Thailand Corresponding author: Chatmongkon Suwannapoom ([email protected]) Academic editor: Truong Nguyen Received: 20 Apr 2021 | Accepted: 25 May 2021 | Published: 28 May 2021 Citation: Lorphengsy S, Nguyen TV, Poyarkov NA, Wu Y-H, Pawangkhanant P, Passorn S, Che J, Suwannapoom C (2021) First national record of Gracixalus quangi Rowley, Dau, Nguyen, Cao & Nguyen, 2011 and G. yunnanensis Yu, Li, Wang, Rao, Wu &Yang, 2019 (Amphibia: Anura: Rhacophoridae) from Thailand. Biodiversity Data Journal 9: e67667. https://doi.org/10.3897/BDJ.9.e67667 Abstract Background The bushfrog genus Gracixalus Delorme, Dubois, Grosjean & Ohler, 2005 is found in southern and south-western China, Vietnam, Laos, Thailand and Myanmar. -

A New Small Montane Species of Philautus (Amphibia: Anura: Rhacophoridae) from Gunung Kinabalu, Sabah, Malaysia (Borneo)
SALAMANDRA 52(2) 77–90 30 June 2016 NewISSN Philautus 0036–3375 from Borneo A new small montane species of Philautus (Amphibia: Anura: Rhacophoridae) from Gunung Kinabalu, Sabah, Malaysia (Borneo) J. Maximilian Dehling1, Masafumi Matsui2 & Paul Yambun Imbun3 1) Institut für Integrierte Naturwissenschaften, Abteilung Biologie, Universität Koblenz-Landau, Universitätsstraße 1, 56070 Koblenz, Germany 2) Graduate School of Human and Environmental Studies, Kyoto University, Sakyo, Kyoto 606–8501, Japan 3) Sabah Parks, P.O. Box 10626, 88806 Kota Kinabalu, Sabah, Malaysia Corresponding author: J. Maximilian Dehling, e-mail: [email protected] Manuscript received: 6 September 2015 Accepted: 17 November 2015 by Alexander Kupfer Abstract. Three populations of small montane bush frogs previously assigned to Philautus mjobergi were compared us- ing morphological, bioacoustic, and genetic (mitochondrial 16S rRNA sequence) data. The comparison revealed that the population from Gunung Kinabalu in Sabah, Malaysia, represents a distinct species that is described herein. Although the new species differs fromP. mjobergi in few morphological characters, most notably the acuminate snout, it is distinguished from this species and all other Bornean congeners by a unique advertisement call and large genetic differences. The popu- lation from Gunung Mulu assigned to P. mjobergi differs from the topotypic population from Gunung Murud in its adver- tisement call and 16S rRNA sequence and probably represents another, undescribed species. Key words. DNA barcoding, advertisement call, Philautus mjobergi, Gunung Mulu, Sarawak, frog. Introduction Smith 1931, Inger 1966 [as P. aurifasciatus], Dring 1987, Malkmus 1989, Inger & Stuebing 1992, Malkmus 1994, Bush frogs of the genus Philautus are represented by 19 Malkmus & Riede 1996, Inger et al. -
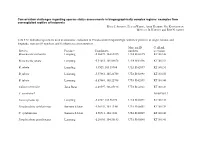
Conservation Challenges Regarding Species Status Assessments in Biogeographically Complex Regions: Examples from Overexploited Reptiles of Indonesia KYLE J
Conservation challenges regarding species status assessments in biogeographically complex regions: examples from overexploited reptiles of Indonesia KYLE J. SHANEY, ELIJAH WOSTL, AMIR HAMIDY, NIA KURNIAWAN MICHAEL B. HARVEY and ERIC N. SMITH TABLE S1 Individual specimens used in taxonomic evaluation of Pseudocalotes tympanistriga, with their province of origin, latitude and longitude, museum ID numbers, and GenBank accession numbers. Museum ID GenBank Species Province Coordinates numbers accession Bronchocela cristatella Lampung -5.36079, 104.63215 UTA R 62895 KT180148 Bronchocela jubata Lampung -5.54653, 105.04678 UTA R 62896 KT180152 B. jubata Lampung -5.5525, 105.18384 UTA R 62897 KT180151 B. jubata Lampung -5.57861, 105.22708 UTA R 62898 KT180150 B. jubata Lampung -5.57861, 105.22708 UTA R 62899 KT180146 Calotes versicolor Jawa Barat -6.49597, 106.85198 UTA R 62861 KT180149 C. versicolor* NC009683.1 Gonocephalus sp. Lampung -5.2787, 104.56198 UTA R 60571 KT180144 Pseudocalotes cybelidermus Sumatra Selatan -4.90149, 104.13401 UTA R 60551 KT180139 P. cybelidermus Sumatra Selatan -4.90711, 104.1348 UTA R 60549 KT180140 Pseudocalotes guttalineatus Lampung -5.28105, 104.56183 UTA R 60540 KT180141 P. guttalineatus Sumatra Selatan -4.90681, 104.13457 UTA R 60501 KT180142 Pseudocalotes rhammanotus Lampung -4.9394, 103.85292 MZB 10804 KT180147 Pseudocalotes species 4 Sumatra Barat -2.04294, 101.31129 MZB 13295 KT211019 Pseudocalotes tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60544 KT180143 P. tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60547 KT180145 Pogona vitticeps* AB166795.1 *Entry to GenBank by previous authors TABLE S2 Reptile species currently believed to occur Java and Sumatra, Indonesia, with IUCN Red List status, and certainty of occurrence. -

The Ecology of Lizard Reproductive Output
Global Ecology and Biogeography, (Global Ecol. Biogeogr.) (2011) ••, ••–•• RESEARCH The ecology of lizard reproductive PAPER outputgeb_700 1..11 Shai Meiri1*, James H. Brown2 and Richard M. Sibly3 1Department of Zoology, Tel Aviv University, ABSTRACT 69978 Tel Aviv, Israel, 2Department of Biology, Aim We provide a new quantitative analysis of lizard reproductive ecology. Com- University of New Mexico, Albuquerque, NM 87131, USA and Santa Fe Institute, 1399 Hyde parative studies of lizard reproduction to date have usually considered life-history Park Road, Santa Fe, NM 87501, USA, 3School components separately. Instead, we examine the rate of production (productivity of Biological Sciences, University of Reading, hereafter) calculated as the total mass of offspring produced in a year. We test ReadingRG6 6AS, UK whether productivity is influenced by proxies of adult mortality rates such as insularity and fossorial habits, by measures of temperature such as environmental and body temperatures, mode of reproduction and activity times, and by environ- mental productivity and diet. We further examine whether low productivity is linked to high extinction risk. Location World-wide. Methods We assembled a database containing 551 lizard species, their phyloge- netic relationships and multiple life history and ecological variables from the lit- erature. We use phylogenetically informed statistical models to estimate the factors related to lizard productivity. Results Some, but not all, predictions of metabolic and life-history theories are supported. When analysed separately, clutch size, relative clutch mass and brood frequency are poorly correlated with body mass, but their product – productivity – is well correlated with mass. The allometry of productivity scales similarly to metabolic rate, suggesting that a constant fraction of assimilated energy is allocated to production irrespective of body size. -

An Updated and Annotated List of Indian Lizards (Reptilia: Sauria) Based on a Review of Distribution Records and Checklists of Indian Reptiles
JoTT REVIEW 2(3): 725-738 An updated and annotated list of Indian lizards (Reptilia: Sauria) based on a review of distribution records and checklists of Indian reptiles P. Dilip Venugopal Department of Entomology, University of Maryland, 4124 Plant Sciences Building, College Park, MD 20742-4454, USA Email: [email protected] Date of publication (online): 26 March 2010 Abstract: Over the past two decades many checklists of reptiles of India and adjacent Date of publication (print): 26 March 2010 countries have been published. These publications have furthered the growth of ISSN 0974-7907 (online) | 0974-7893 (print) knowledge on systematics, distribution and biogeography of Indian reptiles, and the field Editor: Aaron Bauer of herpetology in India in general. However, the reporting format of most such checklists of Indian reptiles does not provide a basis for direct verification of the information presented. Manuscript details: As a result, mistakes in the inclusion and omission of species have been perpetuated Ms # o2083 and the exact number of reptile species reported from India still remains unclear. A Received 21 October 2008 Final received 31 December 2009 verification of the current listings based on distributional records and review of published Finally accepted 14 February 2010 checklists revealed that 199 species of lizards (Reptilia: Sauria) are currently validly reported on the basis of distributional records within the boundaries of India. Seventeen Citation: Venugopal, P.D. (2010). An updated other lizard species have erroneously been included in earlier checklists of Indian reptiles. and annotated list of Indian lizards (Reptilia: Omissions of species by these checklists have been even more numerous than Souria) based on a review of distribution records and checklists of Indian reptiles. -

A Phylogeny and Revised Classification of Squamata, Including 4161 Species of Lizards and Snakes
BMC Evolutionary Biology This Provisional PDF corresponds to the article as it appeared upon acceptance. Fully formatted PDF and full text (HTML) versions will be made available soon. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes BMC Evolutionary Biology 2013, 13:93 doi:10.1186/1471-2148-13-93 Robert Alexander Pyron ([email protected]) Frank T Burbrink ([email protected]) John J Wiens ([email protected]) ISSN 1471-2148 Article type Research article Submission date 30 January 2013 Acceptance date 19 March 2013 Publication date 29 April 2013 Article URL http://www.biomedcentral.com/1471-2148/13/93 Like all articles in BMC journals, this peer-reviewed article can be downloaded, printed and distributed freely for any purposes (see copyright notice below). Articles in BMC journals are listed in PubMed and archived at PubMed Central. For information about publishing your research in BMC journals or any BioMed Central journal, go to http://www.biomedcentral.com/info/authors/ © 2013 Pyron et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes Robert Alexander Pyron 1* * Corresponding author Email: [email protected] Frank T Burbrink 2,3 Email: [email protected] John J Wiens 4 Email: [email protected] 1 Department of Biological Sciences, The George Washington University, 2023 G St. -

Biodiversity and Natural History of Amphibians and Reptiles in Kerinci Seblat National Park, Sumatra, Indonesia (2005, 2006, 2007, 2008)
BIODIVERSITY AND NATURAL HISTORY OF AMPHIBIANS AND REPTILES IN KERINCI SEBLAT NATIONAL PARK, SUMATRA, INDONESIA (2005, 2006, 2007, 2008) Rhacophorus bifasciatus (Photograpg by H. Kurniati) By : HELLEN KURNIATI Research Center for Biology-LIPI, Widyasatwaloka Building-LIPI, Jalan Raya Cibinong km 46, Cibinong 16911, West Java, INDONESIA Phone : +62-21-8765056 Fax : +62-21-8765068 Email : <[email protected]> and <[email protected]> CONTENTS ACKNOWLEDGEMENTS ………………………………………………………………………………. 5 SUMMARY ………………………………………………………………………………………………. 6 INTRODUCTION ……………………………………………………………………............................... 6 METHODS ……………………………………………………………………………………………….. 7 A. Study Areas and Habitats ………………………………………………………............................... 7 A.1. Phase 1 (January-March 2005) ……………………………………………................................... 8 A.2. Phase 2 (February-March 2006) ………………………………………......................................... 11 A.3. Phase 3 (January-March 2007)…………………………………………….................................... 13 A.4. Phase 4 (February 2008) ................................................................................................................. 15 B. Survey Methods ………………………………………………………………….............................. 16 B.1. Lighting ………………………………………………………………………............................. 17 B.2. Hand Collection ………………………………………………………………............................. 17 B.3. Trapping ………………………………………………………………………............................ 17 C. Analysis ………………………………………………………………………….............................. 18 RESULTS