Docking on Lipid II—A Widespread Mechanism for Potent Bactericidal Activities of Antibiotic Peptides
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules As Potent Antibacterial and Antifungal Agents
Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules as Potent Antibacterial and Antifungal Agents Dissertation Zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) Vorgelegt der Naturwissenschaftlichen Fakultät I Institut für Pharmazie Fachbereich für Pharmazeutische Chemie der Martin-Luther-Universität Halle-Wittenberg von Kaveh Yasrebi Geboren am 09.14.1987 in Teheran/Iran (Islamische Republik) Gutachter: 1. Prof. Dr. Andreas Hilgeroth (Martin-Luther-Universität Halle-Wittenberg, Germany) 2. Prof. Dr. Sibel Süzen (Ankara Üniversitesi, Turkey) 3. Prof. Dr. Michael Lalk (Ernst-Moritz-Arndt-Universität Greifswald, Germany) Halle (Saale), den 21. Juli 2020 Selbstständigkeitserklärung Hiermit erkläre ich gemäß § 5 (2) b der Promotionsordnung der Naturwissenschaftlichen Fakultät I – Institut für Pharmazie der Martin-Luther-Universität Halle-Wittenberg, dass ich die vorliegende Arbeit selbstständig und ohne Benutzung anderer als der angegebenen Hilfsmittel und Quellen angefertigt habe. Alle Stellen, die wörtlich oder sinngemäß aus Veröffentlichungen entnommen sind, habe ich als solche kenntlich gemacht. Ich erkläre ferner, dass diese Arbeit in gleicher oder ähnlicher Form bisher keiner anderen Prüfbehörde zur Erlangung des Doktorgrades vorgelegt wurde. Halle (Saale), den 21. Juli 2020 Kaveh Yasrebi Acknowledgement This study was carried out from June 2015 to July 2017 in the Research Group of Drug Development and Analysis led by Prof. Dr. Andreas Hilgeroth at the Institute of Pharmacy, Martin-Luther-Universität Halle-Wittenberg. I would like to thank all the people for their participation who supported my work in this way and helped me obtain good results. First of all, I would like to express my gratitude to Prof. Dr. Andreas Hilgeroth for providing me with opportunity to carry out my Ph.D. -

A New Antibiotic Kills Pathogens Without Detectable Resistance
ARTICLE doi:10.1038/nature14098 A new antibiotic kills pathogens without detectable resistance Losee L. Ling1*, Tanja Schneider2,3*, Aaron J. Peoples1, Amy L. Spoering1, Ina Engels2,3, Brian P. Conlon4, Anna Mueller2,3, Till F. Scha¨berle3,5, Dallas E. Hughes1, Slava Epstein6, Michael Jones7, Linos Lazarides7, Victoria A. Steadman7, Douglas R. Cohen1, Cintia R. Felix1, K. Ashley Fetterman1, William P. Millett1, Anthony G. Nitti1, Ashley M. Zullo1, Chao Chen4 & Kim Lewis4 Antibiotic resistance is spreading faster than the introduction of new compounds into clinical practice, causing a public health crisis. Most antibiotics were produced by screening soil microorganisms, but this limited resource of cultivable bacteria was overmined by the 1960s. Synthetic approaches to produce antibiotics have been unable to replace this platform. Uncultured bacteria make up approximately 99% of all species in external environments, and are an untapped source of new antibiotics. We developed several methods to grow uncultured organisms by cultivation in situ or by using specific growth factors. Here we report a new antibiotic that we term teixobactin, discovered in a screen of uncultured bacteria. Teixobactin inhibits cell wall synthesis by binding to a highly conserved motif of lipid II (precursor of peptidoglycan) and lipid III (precursor of cell wall teichoic acid). We did not obtain any mutants of Staphylococcus aureus or Mycobacterium tuberculosis resistant to teixobactin. The properties of this compound suggest a path towards developing antibiotics that are likely to avoid development of resistance. Widespread introduction of antibiotics in the 1940s, beginning with factors through the chambers enables growth of uncultured bacteria in penicillin1,2 and streptomycin3, transformed medicine, providing effec- their natural environment. -

Investigational Drug Therapies Currently in Early-Stage Clinical Development for the Treatment of Clostridioides (Clostridium) Difficile Infection Mai-Chi N
EXPERT OPINION ON INVESTIGATIONAL DRUGS https://doi.org/10.1080/13543784.2019.1581763 REVIEW Investigational drug therapies currently in early-stage clinical development for the treatment of clostridioides (clostridium) difficile infection Mai-Chi N. Trana,b, Ravina Kullarc and Ellie J. C. Goldsteind,e aDepartment of Pharmacy, Providence St. John’s Health Center, Santa Monica, CA, USA; bDepartment of Pharmacy, Clinica Juan Pablo Medical Group, Los Angeles, CA, USA; cDoctor Evidence, LLC, Santa Monica, CA, USA; dR M Alden Research Laboratory, Santa Monica, CA, USA; eDavid Geffen School of Medicine, Los Angeles, CA, USA ABSTRACT ARTICLE HISTORY Introduction: Clostridioides (Clostridium) difficile Infection (CDI) is an urgent global threat causing Received 16 August 2018 ~500,000 infections annually in the United States of America (USA) and is associated with a 36% 30- Accepted 8 February 2019 day attributable mortality rate. Despite the availability of three therapeutic agents, CDI recurrence KEYWORDS – – occurs in 20 40% of patients, with a 30 40% second recurrence rate in these patients. Consequently, ACX-362F; DS-2969b; there is a need for novel agents for treating CDI. LFF571; ribaxamase; Areas covered: We searched MEDLINE, PubMed, Embase, Web of Science, Cochrane Central Register of ridinilazole; RBX 2660; Controlled Trials, and ClinicalTrials.gov for agents in early stages of clinical development. CRS3123; MCB3681/ These drugs include ACX-362E, DS-2969b, LFF 571, RBX2660, ribaxamase, ridinilazole that have MCB3837 advanced to at least phase 2 and several other drugs in phase 1 development. Expert opinion: The challenge for these new agents is three-fold: (1) to have a novel approach such as a different target/mechanism of action; (2) be ‘significantly’ better than existing agents in regard to ‘sustained clinical response’; or (3) be priced at a reasonable cost when it comes to market or perhaps all three. -

Novel Antimicrobial Agents Inhibiting Lipid II Incorporation Into Peptidoglycan Essay MBB
27 -7-2019 Novel antimicrobial agents inhibiting lipid II incorporation into peptidoglycan Essay MBB Mark Nijland S3265978 Supervisor: Prof. Dr. Dirk-Jan Scheffers Molecular Microbiology University of Groningen Content Abstract..............................................................................................................................................2 1.0 Peptidoglycan biosynthesis of bacteria ........................................................................................3 2.0 Novel antimicrobial agents ...........................................................................................................4 2.1 Teixobactin ...............................................................................................................................4 2.2 tridecaptin A1............................................................................................................................7 2.3 Malacidins ................................................................................................................................8 2.4 Humimycins ..............................................................................................................................9 2.5 LysM ........................................................................................................................................ 10 3.0 Concluding remarks .................................................................................................................... 11 4.0 references ................................................................................................................................. -
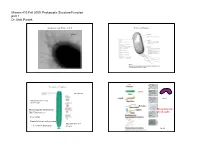
Parsek Micro410 Lecture
Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Organization of the Prokaryotic Cell Prokaryotic Structures fimbriae Size Range of Prokaryotes bacillus See Table 4.1 (rigid) vibrio Nanobacteria 0.05‐0.2 µm (0.14‐0.2 µm) (flexible) Thiomargarita namibiensis Mycoplasma are 700‐750 µm (Fig. 4.2) pleomorphic green alga Nanochlorum eukaryotum Mycoplasma 0.1‐ ~1-2 µm in diameter 0.3 µm Fig. 4.1 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Staining cells for Microscopic observation Cell Arrangements Motility- ~80% of prokaryotes are motile streptococcus Staining properties: Gram Stain staphylococcus Fig. 2.3 Gram Stain (1884) (Bacteria) Gram-negative mixed culture Gram-positive Fig. 2.3 and 2.4 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Functions of the cytoplasmic membrane The phospholipid bi‐layer Fig. 4.9 Fig. 4.4 What is the structure of bacterial phospholipids? Other components of the cytoplasmic membrane Figs. 4.5‐4.6 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Archaeal membranes can be a lipid monolayer Archaeal phospholipids have an ether linkage Fig. 4.7 Fig. 4.8 Importance of Cell Wall Schematic diagram cell wall • Provides rigidity to cell allowing cell to withstand the large osmotic/ionic Fig. 4.16 changes a bacterium may experience in its environment, and turgor pressure of cytoplasm (conc. of solutes in cytoplasm). Cell lysis • May have a role in shape determination. • Provides a barrier against certain toxic chemical and biological agents. • Site of action of some of the most commonly used antibiotics used to treat bacterial infections (penicillin family). -
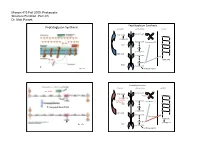
Parsek Lecture #3
Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Peptidoglycan Synthesis Peptidoglycan Synthesis cytoplasm cell membrane cell wall Bactoprenol-P Pi UDP-NAM M G pentapeptide G M Bactoprenol Bactoprenol-P-P P M UMP G P NAM G M pentapeptide M G UDP-NAG Bactoprenol G P NAM‐NAG P NAM-NAG UMP pentapeptide Fig. 6.7a Interbridge peptide Peptidoglycan Synthesis Cross-linking of Peptidoglycan Strands cytoplasm cell membrane cell wall autolysins Bactoprenol-P Pi UDP-NAM Bacitracin M G pentapeptide G D-cycloserine Bactoprenol M (Oxamycin) Bactoprenol-P-P P M UMP G P Transpeptidase (FtsI) NAM G M pentapeptide M G UDP-NAG Bactoprenol G Vancomycin P NAM‐NAG P NAM-NAG pentapeptide UMP Fig. 6.7b pentapeptide Interbridge peptide Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cross-linking of Peptidoglycan Strands Antibiotic Resistance autolysins • Inactivate antibiotic β-lactamase (penicillinase) Clavulanic acid β-lactams Augmentin and Trimentin (combination of clavulanic acid and transpeptidase amoxicillin or ampicillin respectively) penicillins and cephalosporins lysozyme • Change chemistry of target site • Limit access of the antibiotic to target site Fig. 6.5 Cell Shape Determination • Modifications made to Peptidoglycan: ‐ lysozyme: Protoplasts/spheroplasts ‐ autolysins Bacillus subtilis ‐ endopeptidase Heliobacter pylori • Protein(s) may play a major role ‐ MreB protein Caulobacter crescentus ‐ MreB has homology to actin, a component of the cytoskeleton of eukaryotes. Shape determining protein‐ crescentin Fig. 6.4 Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cell Wall Gram-positive Bacteria intermediate filaments in the bacteria Caulobacter crescentus glycerol similar predicted structures of crescentin and intermediate filaments Fig. -

Abstract a Practical Synthesis of N -Fmoc Protected L-Threo- -Hydroxyaspartic Acid Derivatives for Coupling
CYCLIC LIPODEPSIPEPTIDES AS LEAD STRUCTURES FOR THE DISCOVERY OF NEW ANTIBIOTICS by Nina Bionda A Dissertation Submitted to the Faculty of The Charles E. Schmidt College of Science in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Florida Atlantic University Boca Raton, FL May 2013 ACKNOWLEDGEMENTS First and foremost, I want to thank my advisor, Dr. Predrag Cudic, for his guidance throughout my PhD endeavors, the endless discussions about science and life in general, and for teaching me to give my best every step of the way. You have been instrumental in my scientific education and for that I am forever in your debt. Immense gratitude goes to Dr. Lepore for embracing the difficult role of a co-advisor and fulfilling it truly beyond what I have expected. I will always appreciate your advice. I also wish to express my heartfelt thanks to my dissertation committee members, Dr. Laszlo Otvos Jr., Dr. Stefan Vetter and Dr. Andrew C. Terentis, for their valuable time and thoughtful suggestions, comments and critiques. I would also like to extend my appreciation to all of the faculty and students of the Department of Chemistry and Biochemistry that I have had contact with during my PhD studies. To Dr. Richard Houghten and everyone at the Torrey Pines Institute for Molecular Studies, thank you for providing me with a “home” for the last two and a half years of my PhD. And special thanks to Dr. Maré Cudic for sharing her scientific experience and the simple things of the everyday life. To Dr. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

Microorganism Solutions
C297-E086 Microorganism Species Analysis and Component Analysis – Detection and Identification of Microorganisms and Metabolite Analysis – Shimadzu’s Microorganism Solutions JQA-0376 Founded in 1875, Shimadzu Corporation, a leader in the development of advanced technologies, has a distinguished history of innovation built on the foundation of contributing to society through science and technology. We maintain a global network of sales, service, technical support and applications centers on six continents, and have established long-term relationships with a host of highly trained distributors located in over 100 countries. For information about Shimadzu, and to contact your local office, please visit our Web site at www.shimadzu.com Microorganism SHIMADZU CORPORATION. International Marketing Division 3. Kanda-Nishikicho 1-chome, Chiyoda-ku, Tokyo 101-8448, Japan Solutions Phone: 81(3)3219-5641 Fax. 81(3)3219-5710 URL http://www.shimadzu.com The contents of this brochure are subject to change without notice. AnalysisAnalysis Printed in Japan 3295-12904-15A-NS TotalTotal SolutionsSolutions forfor MicroorganismMicroorganism AnalysisAnalysis andand TestingTesting Shimadzu Supports Everybody Working with Microorganisms While we cannot easily see microorganisms, they are closely linked to our lives. From ancient times, microorganisms have expanded Microorganisms are researched and utilized for a wide range of purposes in many research and industry fields. the human diet by providing fermented foods including alcohol and pickles. In recent years, microorganisms have become The major purposes are inspection and control, identification, searching for new microorganisms, functional investigations, and indispensable in the production of useful substances, such as antibiotics, taste components, and vitamins. Microorganisms are also industrial applications. essential for environmental sustainability and improvements, including wastewater treatment, soil cleanup, and atmospheric balance. -

Effect of Msa on Antibiotic Resistance and Allelic Replacement of Pkor1 in Staphylococcus Aureus
The University of Southern Mississippi The Aquila Digital Community Honors Theses Honors College Fall 12-2012 Effect of msa on Antibiotic Resistance and Allelic Replacement of pKOR1 in Staphylococcus aureus Jordan M. Towne University of Southern Mississippi Follow this and additional works at: https://aquila.usm.edu/honors_theses Recommended Citation Towne, Jordan M., "Effect of msa on Antibiotic Resistance and Allelic Replacement of pKOR1 in Staphylococcus aureus" (2012). Honors Theses. 103. https://aquila.usm.edu/honors_theses/103 This Honors College Thesis is brought to you for free and open access by the Honors College at The Aquila Digital Community. It has been accepted for inclusion in Honors Theses by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. The University of Southern Mississippi Effect of msa on antibiotic resistance and allelic replacement of pKOR1 in Staphylococcus aureus by Jordan Towne A Thesis Submitted to the Honors College of The University of Southern Mississippi in Partial Fulfillment of the Requirements for the Degree of Bachelor of Science in the Department of Biological Sciences December 2012 ii Approved By: ____________________________ Mohamed O. Elasri Department of Biological Sciences ____________________________ Glenmore Shearer, Chair Department of Biological Sciences ___________________________ David R. Davies, Dean Honors College iii Abstract Staphylococcus aureus is an important human pathogen that causes hospital and community-acquired infections (52). These infections are difficult to treat due to resistance to a wide range of antibiotics and spread of antibiotic-resistant strains (13, 52). S. aureus causes infection by regulation of accessory genes encoding for expression of factors contributing to virulence (9, 11, 12, 29, 34, 43, 45), including severe infection, biofilm formation, autolysis, and antibiotic resistance (4, 5, 7, 27, 56). -

Escherichia Coli K–12 Derived from the Ecocyc Database Daniel S Weaver1*,Ingridmkeseler1,Amandamackie2, Ian T Paulsen2 and Peter D Karp1
Weaver et al. BMC Systems Biology 2014, 8:79 http://www.biomedcentral.com/1752-0509/8/79 RESEARCH ARTICLE OpenAccess A genome-scale metabolic flux model of Escherichia coli K–12 derived from the EcoCyc database Daniel S Weaver1*,IngridMKeseler1,AmandaMackie2, Ian T Paulsen2 and Peter D Karp1 Abstract Background: Constraint-based models of Escherichia coli metabolic flux have played a key role in computational studies of cellular metabolism at the genome scale. We sought to develop a next-generation constraint-based E. coli model that achieved improved phenotypic prediction accuracy while being frequently updated and easy to use. We also sought to compare model predictions with experimental data to highlight open questions in E. coli biology. Results: We present EcoCyc–18.0–GEM, a genome-scale model of the E. coli K–12 MG1655 metabolic network. The model is automatically generated from the current state of EcoCyc using the MetaFlux software, enabling the release of multiple model updates per year. EcoCyc–18.0–GEM encompasses 1445 genes, 2286 unique metabolic reactions, and 1453 unique metabolites. We demonstrate a three-part validation of the model that breaks new ground in breadth and accuracy: (i) Comparison of simulated growth in aerobic and anaerobic glucose culture with experimental results from chemostat culture and simulation results from the E. coli modeling literature. (ii) Essentiality prediction for the 1445 genes represented in the model, in which EcoCyc–18.0–GEM achieves an improved accuracy of 95.2% in predicting the growth phenotype of experimental gene knockouts. (iii) Nutrient utilization predictions under 431 different media conditions, for which the model achieves an overall accuracy of 80.7%. -

The Heme Sensing Response Regulator Hssr in Staphylococcus Aureus
Thomsen et al. BMC Microbiology 2010, 10:307 http://www.biomedcentral.com/1471-2180/10/307 RESEARCH ARTICLE Open Access The heme sensing response regulator HssR in Staphylococcus aureus but not the homologous RR23 in Listeria monocytogenes modulates susceptibility to the antimicrobial peptide plectasin Line E Thomsen1*, Caroline T Gottlieb2,5, Sanne Gottschalk1, Tim T Wodskou3, Hans-Henrik Kristensen4, Lone Gram2, Hanne Ingmer1 Abstract Background: Host defence peptides (HDPs), also known as antimicrobial peptides (AMPs), have emerged as potential new therapeutics and their antimicrobial spectrum covers a wide range of target organisms. However, the mode of action and the genetics behind the bacterial response to HDPs is incompletely understood and such knowledge is required to evaluate their potential as antimicrobial therapeutics. Plectasin is a recently discovered HDP active against Gram-positive bacteria with the human pathogen, Staphylococcus aureus (S. aureus) being highly susceptible and the food borne pathogen, Listeria monocytogenes (L. monocytogenes) being less sensitive. In the present study we aimed to use transposon mutagenesis to determine the genetic basis for S. aureus and L. monocytogenes susceptibility to plectasin. Results: In order to identify genes that provide susceptibility to plectasin we constructed bacterial transposon mutant libraries of S. aureus NCTC8325-4 and L. monocytogenes 4446 and screened for increased resistance to the peptide. No resistant mutants arose when L. monocytogenes was screened on plates containing 5 and 10 fold Minimal Inhibitory Concentration (MIC) of plectasin. However, in S. aureus, four mutants with insertion in the heme response regulator (hssR) were 2-4 fold more resistant to plectasin as compared to the wild type.