Detection of Histoplasma DNA from Tissue Blocks by a Specific
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Congo DRC, Kamwiziku. Mycoses 2021
Received: 8 April 2021 | Revised: 1 June 2021 | Accepted: 10 June 2021 DOI: 10.1111/myc.13339 REVIEW ARTICLE Serious fungal diseases in Democratic Republic of Congo – Incidence and prevalence estimates Guyguy K. Kamwiziku1 | Jean- Claude C. Makangara1 | Emma Orefuwa2 | David W. Denning2,3 1Department of Microbiology, Kinshasa University Hospital, University of Abstract Kinshasa, Kinshasa, Democratic Republic A literature review was conducted to assess the burden of serious fungal infections of Congo 2Global Action Fund for Fungal Infections, in the Democratic Republic of the Congo (DRC) (population 95,326,000). English and Geneva, Switzerland French publications were listed and analysed using PubMed/Medline, Google Scholar 3 Manchester Fungal Infection Group, The and the African Journals database. Publication dates spanning 1943– 2020 were in- University of Manchester, Manchester Academic Health Science Centre, cluded in the scope of the review. From the analysis of published articles, we estimate Manchester, UK a total of about 5,177,000 people (5.4%) suffer from serious fungal infections in the Correspondence DRC annually. The incidence of cryptococcal meningitis, Pneumocystis jirovecii pneu- Guyguy K. Kamwiziku, Department monia in adults and invasive aspergillosis in AIDS patients was estimated at 6168, of Microbiology, Kinshasa University Hospital, University of Kinshasa, Congo. 2800 and 380 cases per year. Oral and oesophageal candidiasis represent 50,470 Email: [email protected] and 28,800 HIV- infected patients respectively. Chronic pulmonary aspergillosis post- tuberculosis incidence and prevalence was estimated to be 54,700. Fungal asthma (allergic bronchopulmonary aspergillosis and severe asthma with fungal sensitiza- tion) probably has a prevalence of 88,800 and 117,200. -

Estimated Burden of Serious Fungal Infections in Ghana
Journal of Fungi Article Estimated Burden of Serious Fungal Infections in Ghana Bright K. Ocansey 1, George A. Pesewu 2,*, Francis S. Codjoe 2, Samuel Osei-Djarbeng 3, Patrick K. Feglo 4 and David W. Denning 5 1 Laboratory Unit, New Hope Specialist Hospital, Aflao 00233, Ghana; [email protected] 2 Department of Medical Laboratory Sciences, School of Biomedical and Allied Health Sciences, College of Health Sciences, University of Ghana, P.O. Box KB-143, Korle-Bu, Accra 00233, Ghana; [email protected] 3 Department of Pharmaceutical Sciences, Faculty of Health Sciences, Kumasi Technical University, P.O. Box 854, Kumasi 00233, Ghana; [email protected] 4 Department of Clinical Microbiology, School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi 00233, Ghana; [email protected] 5 National Aspergillosis Centre, Wythenshawe Hospital and the University of Manchester, Manchester M23 9LT, UK; [email protected] * Correspondence: [email protected] or [email protected] or [email protected]; Tel.: +233-277-301-300; Fax: +233-240-190-737 Received: 5 March 2019; Accepted: 14 April 2019; Published: 11 May 2019 Abstract: Fungal infections are increasingly becoming common and yet often neglected in developing countries. Information on the burden of these infections is important for improved patient outcomes. The burden of serious fungal infections in Ghana is unknown. We aimed to estimate this burden. Using local, regional, or global data and estimates of population and at-risk groups, deterministic modelling was employed to estimate national incidence or prevalence. Our study revealed that about 4% of Ghanaians suffer from serious fungal infections yearly, with over 35,000 affected by life-threatening invasive fungal infections. -

Serious Fungal Infections in Peru
Eur J Clin Microbiol Infect Dis DOI 10.1007/s10096-017-2924-9 ORIGINAL ARTICLE Serious fungal infections in Peru B. Bustamante1 & D. W. Denning2 & P. E. Campos3 Received: 21 December 2016 /Accepted: 21 December 2016 # Springer-Verlag Berlin Heidelberg 2017 Abstract Epidemiological data about mycotic diseases are This first attempt to assess the fungal burden in Peru needs limited in Peru and estimation of the fungal burden has not to be refined. We believe the figure obtained is an underesti- been previously attempted. Data were obtained from the mation, because of under diagnosis, non-mandatory reporting Peruvian National Institute of Statistics and Informatics, and lack of a surveillance system and of good data about the UNAIDS and from the Ministry of Health’s publications. size of populations at risk. We also searched the bibliography for Peruvian data on my- cotic diseases, asthma, COPD, cancer and transplants. Incidence or prevalence for each fungal disease were estimat- Introduction ed in specific populations at risk. The Peruvian population for 2015 was 31,151,543. In 2014, the estimated number of HIV/ While candidemia and invasive aspergillosis (IA) are clearly AIDS and pulmonary tuberculosis cases was 88,625, 38,581 recognized as important causes of morbidity and mortality, of them not on ART, and 22,027, respectively. A total of other mycotic agents or different clinical presentations are im- 581,174 cases of fungal diseases were estimated, representing portant in specific regions. Socio-economic and geo- approximately 1.9% of the Peruvian population. This figure ecological characteristics and size of susceptible populations includes 498,606, 17,361 and 4,431 vulvovaginal, oral and are the main determinants of variations on incidence of fungal esophageal candidiasis, respectively, 1,557 candidemia cases, infections across the world. -

A Case of Disseminated Histoplasmosis Detected in Peripheral Blood Smear Staining Revealing AIDS at Terminal Phase in a Female Patient from Cameroon
Hindawi Publishing Corporation Case Reports in Medicine Volume 2012, Article ID 215207, 3 pages doi:10.1155/2012/215207 Case Report A Case of Disseminated Histoplasmosis Detected in Peripheral Blood Smear Staining Revealing AIDS at Terminal Phase in a Female Patient from Cameroon Christine Mandengue Ebenye Cliniques Universitaires des Montagnes, Bangangt´e, Cameroon Correspondence should be addressed to Christine Mandengue Ebenye, [email protected] Received 28 August 2012; Revised 2 November 2012; Accepted 5 November 2012 Academic Editor: Ingo W. Husstedt Copyright © 2012 Christine Mandengue Ebenye. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Histoplasmosis is endemic in the American continent and also in Sub-Saharan Africa, coexisting with the African histoplasmosis. Immunosuppressed patients, especially those with advanced HIV infection develop a severe disseminated histoplasmosis with fatal prognosis. The definitive diagnosis of disseminated histoplasmosis is based on the detection of Histoplasma capsulatum from patient’ tissues samples or body fluids. Among the diagnostic tests peripheral blood smear staining is not commonly used. Nonetheless a few publications reveal that Histoplasma capsulatum has been discovered by chance using this method in HIV infected patients with chronic fever and hence revealed AIDS at the terminal phase. We report a new case detected in a Cameroonian woman without any previous history of HIV infection. Peripheral blood smear staining should be commonly used for the diagnosis of disseminated histoplasmosis in the Sub-Saharan Africa, where facilities for mycology laboratories are unavailable. 1. Introduction 1987 [2]. -

Review Article Fungal Dimorphism and Virulence: Molecular Mechanisms for Temperature Adaptation, Immune Evasion, and in Vivo Survival
Hindawi Mediators of Inflammation Volume 2017, Article ID 8491383, 8 pages https://doi.org/10.1155/2017/8491383 Review Article Fungal Dimorphism and Virulence: Molecular Mechanisms for Temperature Adaptation, Immune Evasion, and In Vivo Survival Gregory M. Gauthier Department of Medicine, Division of Infectious Diseases, University of Wisconsin School of Medicine & Public Health, Madison, WI, USA Correspondence should be addressed to Gregory M. Gauthier; [email protected] Received 18 November 2016; Accepted 12 April 2017; Published 23 May 2017 Academic Editor: Anamélia L. Bocca Copyright © 2017 Gregory M. Gauthier. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The thermally dimorphic fungi are a unique group of fungi within the Ascomycota phylum that respond to shifts in temperature by ° ° converting between hyphae (22–25 C) and yeast (37 C). This morphologic switch, known as the phase transition, defines the biology and lifestyle of these fungi. The conversion to yeast within healthy and immunocompromised mammalian hosts is essential for virulence. In the yeast phase, the thermally dimorphic fungi upregulate genes involved with subverting host immune defenses. This review highlights the molecular mechanisms governing the phase transition and recent advances in how the phase transition promotes infection. 1. Introduction are more typically phytopathogenic or entomopathogenic. For example, Ophiostoma novo-ulmi, the etiologic agent The ability for fungi to switch between different morphologic of Dutch elm disease, has destroyed millions of elm trees forms is widespread throughout the fungal kingdom and is a in Europe and United States [13]. -

Series Fungal Infections 8 Improvement of Fungal Disease
Series Fungal infections 8 Improvement of fungal disease identification and management: combined health systems and public health approaches Donald C Cole, Nelesh P Govender, Arunaloke Chakrabarti, Jahit Sacarlal, David W Denning More than 1·6 million people are estimated to die of fungal diseases each year, and about a billion people have Lancet Infect Dis 2017 cutaneous fungal infections. Fungal disease diagnosis requires a high level of clinical suspicion and specialised Published Online laboratory testing, in addition to culture, histopathology, and imaging expertise. Physicians with varied specialist July 31, 2017 training might see patients with fungal disease, yet it might remain unrecognised. Antifungal treatment is more http://dx.doi.org/10.1016 S1473-3099(17)30308-0 complex than treatment for bacterial or most viral infections, and drug interactions are particularly problematic. See Online/Series Health systems linking diagnostic facilities with therapeutic expertise are typically fragmented, with major elements http://dx.doi.org/10.1016/ missing in thousands of secondary care and hospital settings globally. In this paper, the last in a Series of eight papers, S1473-3099(17)30303-1, we describe these limitations and share responses involving a combined health systems and public health framework http://dx.doi.org/10.1016/ illustrated through country examples from Mozambique, Kenya, India, and South Africa. We suggest a mainstreaming S1473-3099(17)30304-3, http://dx.doi.org/10.1016/ approach including greater integration of fungal diseases into existing HIV infection, tuberculosis infection, diabetes, S1473-3099(17)30309-2, chronic respiratory disease, and blindness health programmes; provision of enhanced laboratory capacity to detect http://dx.doi.org/10.1016/ fungal diseases with associated surveillance systems; procurement and distribution of low-cost, high-quality antifungal S1473-3099(17)30306-7, medicines; and concomitant integration of fungal disease into training of the health workforce. -

Severe Chromoblastomycosis-Like Cutaneous Infection Caused by Chrysosporium Keratinophilum
fmicb-08-00083 January 25, 2017 Time: 11:0 # 1 CASE REPORT published: 25 January 2017 doi: 10.3389/fmicb.2017.00083 Severe Chromoblastomycosis-Like Cutaneous Infection Caused by Chrysosporium keratinophilum Juhaer Mijiti1†, Bo Pan2,3†, Sybren de Hoog4, Yoshikazu Horie5, Tetsuhiro Matsuzawa6, Yilixiati Yilifan1, Yong Liu1, Parida Abliz7, Weihua Pan2,3, Danqi Deng8, Yun Guo8, Peiliang Zhang8, Wanqing Liao2,3* and Shuwen Deng2,3,7* 1 Department of Dermatology, People’s Hospital of Xinjiang Uygur Autonomous Region, Urumqi, China, 2 Department of Dermatology, Shanghai Changzheng Hospital, Second Military Medical University, Shanghai, China, 3 Key Laboratory of Molecular Medical Mycology, Shanghai Changzheng Hospital, Second Military Medical University, Shanghai, China, 4 CBS-KNAW Fungal Biodiversity Centre, Royal Netherlands Academy of Arts and Sciences, Utrecht, Netherlands, 5 Medical Mycology Research Center, Chiba University, Chiba, Japan, 6 Department of Nutrition Science, University of Nagasaki, Nagasaki, Japan, 7 Department of Dermatology, First Hospital of Xinjiang Medical University, Urumqi, China, 8 Department of Dermatology, The Second Affiliated Hospital of Kunming Medical University, Kunming, China Chrysosporium species are saprophytic filamentous fungi commonly found in the Edited by: soil, dung, and animal fur. Subcutaneous infection caused by this organism is Leonard Peruski, rare in humans. We report a case of subcutaneous fungal infection caused by US Centers for Disease Control and Prevention, USA Chrysosporium keratinophilum in a 38-year-old woman. The patient presented with Reviewed by: severe chromoblastomycosis-like lesions on the left side of the jaw and neck for 6 years. Nasib Singh, She also got tinea corporis on her trunk since she was 10 years old. -

Managing Athlete's Foot
South African Family Practice 2018; 60(5):37-41 S Afr Fam Pract Open Access article distributed under the terms of the ISSN 2078-6190 EISSN 2078-6204 Creative Commons License [CC BY-NC-ND 4.0] © 2018 The Author(s) http://creativecommons.org/licenses/by-nc-nd/4.0 REVIEW Managing athlete’s foot Nkatoko Freddy Makola,1 Nicholus Malesela Magongwa,1 Boikgantsho Matsaung,1 Gustav Schellack,2 Natalie Schellack3 1 Academic interns, School of Pharmacy, Sefako Makgatho Health Sciences University 2 Clinical research professional, pharmaceutical industry 3 Professor, School of Pharmacy, Sefako Makgatho Health Sciences University *Corresponding author, email: [email protected] Abstract This article is aimed at providing a succinct overview of the condition tinea pedis, commonly referred to as athlete’s foot. Tinea pedis is a very common fungal infection that affects a significantly large number of people globally. The presentation of tinea pedis can vary based on the different clinical forms of the condition. The symptoms of tinea pedis may range from asymptomatic, to mild- to-severe forms of pain, itchiness, difficulty walking and other debilitating symptoms. There is a range of precautionary measures available to prevent infection, and both oral and topical drugs can be used for treating tinea pedis. This article briefly highlights what athlete’s foot is, the different causes and how they present, the prevalence of the condition, the variety of diagnostic methods available, and the pharmacological and non-pharmacological management of the -

Oral Antifungals Month/Year of Review: July 2015 Date of Last
© Copyright 2012 Oregon State University. All Rights Reserved Drug Use Research & Management Program Oregon State University, 500 Summer Street NE, E35 Salem, Oregon 97301-1079 Phone 503-947-5220 | Fax 503-947-1119 Class Update with New Drug Evaluation: Oral Antifungals Month/Year of Review: July 2015 Date of Last Review: March 2013 New Drug: isavuconazole (a.k.a. isavunconazonium sulfate) Brand Name (Manufacturer): Cresemba™ (Astellas Pharma US, Inc.) Current Status of PDL Class: See Appendix 1. Dossier Received: Yes1 Research Questions: Is there any new evidence of effectiveness or safety for oral antifungals since the last review that would change current PDL or prior authorization recommendations? Is there evidence of superior clinical cure rates or morbidity rates for invasive aspergillosis and invasive mucormycosis for isavuconazole over currently available oral antifungals? Is there evidence of superior safety or tolerability of isavuconazole over currently available oral antifungals? • Is there evidence of superior effectiveness or safety of isavuconazole for invasive aspergillosis and invasive mucormycosis in specific subpopulations? Conclusions: There is low level evidence that griseofulvin has lower mycological cure rates and higher relapse rates than terbinafine and itraconazole for adult 1 onychomycosis.2 There is high level evidence that terbinafine has more complete cure rates than itraconazole (55% vs. 26%) for adult onychomycosis caused by dermatophyte with similar discontinuation rates for both drugs.2 There is low -

Therapeutic Class Overview Onychomycosis Agents
Therapeutic Class Overview Onychomycosis Agents Therapeutic Class • Overview/Summary: This review will focus on the antifungal agents Food and Drug Administration (FDA)-approved for the treatment of onychomycosis.1-9 Onychomycosis is a progressive infection of the nail bed which may extend into the matrix or plate, leading to destruction, deformity, thickening and discoloration. Of note, these agents are only indicated when specific types of fungus have caused the infection, and are listed in Table 1. Additionally, ciclopirox is only FDA-approved for mild to moderate onychomycosis without lunula involvement.1 The mechanisms by which these agents exhibit their antifungal effects are varied. For ciclopirox (Penlac®) the exact mechanism is unknown. It is believed to block fungal transmembrane transport, causing intracellular depletion of essential substrates and/or ions and to interfere with ribonucleic acid (RNA) and deoxyribonucleic acid (DNA).1 The azole antifungals, efinaconazole (Jublia®) and itraconazole tablets (Onmel®) and capsules (Sporanox®) works via inhibition of fungal lanosterol 14-alpha-demethylase, an enzyme necessary for the biosynthesis of ergosterol. By decreasing ergosterol concentrations, the fungal cell membrane permeability is increased, which results in leakage of cellular contents.2,5,6 Griseofulvin microsize (Grifulvin V®) and ultramicrosize (GRIS-PEG®) disrupts the mitotic spindle, arresting metaphase of cell division. Griseofulvin may also produce defective DNA that is unable to replicate. The ultramicrosize tablets are absorbed from the gastrointestinal tract at approximately one and one-half times that of microsize griseofulvin, which allows for a lower dose of griseofulvin to be administered.3,4 Tavaborole (Kerydin®), is an oxaborole antifungal that interferes with protein biosynthesis by inhibiting leucyl-transfer ribonucleic acid (tRNA) synthase (LeuRS), which prevents translation of tRNA by LeuRS.7 The final agent used for the treatment of onychomycosis, terbinafine hydrochloride (Lamisil®), is an allylamine antifungal. -

Pulmonary Aspergillosis: What CT Can Offer Before Radiology Section It Is Too Late!
DOI: 10.7860/JCDR/2016/17141.7684 Review Article Pulmonary Aspergillosis: What CT can Offer Before Radiology Section it is too Late! AKHILA PRASAD1, KSHITIJ AGARWAL2, DESH DEEPAK3, SWAPNDEEP SINGH ATWAL4 ABSTRACT Aspergillus is a large genus of saprophytic fungi which are present everywhere in the environment. However, in persons with underlying weakened immune response this innocent bystander can cause fatal illness if timely diagnosis and management is not done. Chest infection is the most common infection caused by Aspergillus in human beings. Radiological investigations particularly Computed Tomography (CT) provides the easiest, rapid and decision making information where tissue diagnosis and culture may be difficult and time-consuming. This article explores the crucial role of CT and offers a bird’s eye view of all the radiological patterns encountered in pulmonary aspergillosis viewed in the context of the immune derangement associated with it. Keywords: Air-crescent, Fungal ball, Halo sign, Invasive aspergillosis INTRODUCTION diagnostic pitfalls one encounters and also addresses the crucial The genus Aspergillus comprises of hundreds of fungal species issue as to when to order for the CT. ubiquitously present in nature; predominantly in the soil and The spectrum of disease that results from the Aspergilla becoming decaying vegetation. Nearly, 60 species of Aspergillus are a resident in the lung is known as ‘Pulmonary Aspergillosis’. An medically significant, owing to their ability to cause infections inert colonization of pulmonary cavities like in cases of tuberculosis in human beings affecting multiple organ systems, chiefly the and Sarcoidosis, where cavity formation is quite common, makes lungs, paranasal sinuses, central nervous system, ears and skin. -
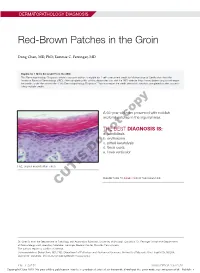
Red-Brown Patches in the Groin
DERMATOPATHOLOGY DIAGNOSIS Red-Brown Patches in the Groin Dong Chen, MD, PhD; Tammie C. Ferringer, MD Eligible for 1 MOC SA Credit From the ABD This Dermatopathology Diagnosis article in our print edition is eligible for 1 self-assessment credit for Maintenance of Certification from the American Board of Dermatology (ABD). After completing this activity, diplomates can visit the ABD website (http://www.abderm.org) to self-report the credits under the activity title “Cutis Dermatopathology Diagnosis.” You may report the credit after each activity is completed or after accumu- lating multiple credits. A 66-year-old man presented with reddish arciform patchescopy in the inguinal area. THE BEST DIAGNOSIS IS: a. candidiasis b. noterythrasma c. pitted keratolysis d. tinea cruris Doe. tinea versicolor H&E, original magnification ×600. PLEASE TURN TO PAGE 419 FOR THE DIAGNOSIS CUTIS Dr. Chen is from the Department of Pathology and Anatomical Sciences, University of Missouri, Columbia. Dr. Ferringer is from the Departments of Dermatology and Laboratory Medicine, Geisinger Medical Center, Danville, Pennsylvania. The authors report no conflict of interest. Correspondence: Dong Chen, MD, PhD, Department of Pathology and Anatomical Sciences, University of Missouri, One Hospital Dr, MA204, DC018.00, Columbia, MO 65212 ([email protected]). 416 I CUTIS® WWW.MDEDGE.COM/CUTIS Copyright Cutis 2018. No part of this publication may be reproduced, stored, or transmitted without the prior written permission of the Publisher. DERMATOPATHOLOGY DIAGNOSIS DISCUSSION THE DIAGNOSIS: Erythrasma rythrasma usually involves intertriginous areas surface (Figure 1) compared to dermatophyte hyphae that (eg, axillae, groin, inframammary area). Patients tend to be parallel to the surface.2 E present with well-demarcated, minimally scaly, red- Pitted keratolysis is a superficial bacterial infection brown patches.