Deletion of Stk11 and Fos in Mouse BLA Projection Neurons Alters Intrinsic 3 Excitability and Impairs Formation of Long-Term Aversive Memory
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Deletion of Stk11 and Fos in Mouse BLA Projection Neurons
RESEARCH ARTICLE Deletion of Stk11 and Fos in mouse BLA projection neurons alters intrinsic excitability and impairs formation of long- term aversive memory David Levitan1†*, Chenghao Liu1†, Tracy Yang1, Yasuyuki Shima1, Jian-You Lin2,3, Joseph Wachutka2, Yasmin Marrero2, Ramin Ali Marandi Ghoddousi1, Eduardo da Veiga Beltrame2, Troy A Richter1, Donald B Katz2,3, Sacha B Nelson1,3* 1Departments of Biology, Brandeis University, Waltham, United States; 2Departments of Psychology, Brandeis University, Waltham, United States; 3Volen Center for Complex Systems, Brandeis University, Waltham, United States Abstract Conditioned taste aversion (CTA) is a form of one-trial learning dependent on basolateral amygdala projection neurons (BLApn). Its underlying cellular and molecular mechanisms remain poorly understood. RNAseq from BLApn identified changes in multiple candidate learning- related transcripts including the expected immediate early gene Fos and Stk11, a master kinase of the AMP-related kinase pathway with important roles in growth, metabolism and development, but not previously implicated in learning. Deletion of Stk11 in BLApn blocked memory prior to training, but not following it and increased neuronal excitability. Conversely, BLApn had reduced excitability following CTA. BLApn knockout of a second learning-related gene, Fos, also increased excitability and impaired learning. Independently increasing BLApn excitability chemogenetically during CTA *For correspondence: also impaired memory. STK11 and C-FOS activation were independent -

An Emerging Acral Melanoma Oncogene
www.impactjournals.com/oncotarget/ Oncotarget, Advance Publications 2011 NUAK2: an emerging acral melanoma oncogene Takeshi Namiki1,2, Sergio G. Coelho1, Vincent J. Hearing1 1 Laboratory of Cell Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20814, USA 2 Department of Dermatology, Yokohama Minato Red Cross Hospital, Yokohama, Kanagawa 231-0801, Japan Correspondence to: Dr. Takeshi Namiki, email: [email protected] Keywords: NUAK2, acral melanoma, migration, metastasis, oncogene Received: September 9, 2011, Accepted: September 10, 2011, Published: September 10, 2011 Copyright: © Namiki et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: Recent technological advances in cancer genomics make it possible to dissect complicated genomic aberrations of melanomas. In particular, several specific genomic aberrations including 11q13 amplification and KIT aberrations have been identified in acral melanomas. We recently identified NUAK2 at 1q32 as a promising oncogene in acral melanomas and reported its significant roles in tumorigenesis in melanoma cells using both in vitro and in vivo analyses. NUAK2 as a member of the AMPK family has several intriguing aspects both as an oncogene and as a tumor suppressor gene. Here we review genomic aberrations of melanomas focusing on acral melanomas to emphasize the possible roles -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5. -

Anti-STK11 / LKB1 Antibody (ARG59116)
Product datasheet [email protected] ARG59116 Package: 50 μg anti-STK11 / LKB1 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes STK11 / LKB1 Tested Reactivity Hu Tested Application WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name STK11 / LKB1 Antigen Species Human Immunogen Recombinant protein corresponding to K62-C430 of Human STK11 / LKB1. Conjugation Un-conjugated Alternate Names hLKB1; Serine/threonine-protein kinase STK11; Liver kinase B1; Renal carcinoma antigen NY-REN-19; LKB1; EC 2.7.11.1; PJS Application Instructions Application table Application Dilution WB 0.1 - 0.5 µg/ml Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 49 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer 0.9% NaCl, 0.2% Na2HPO4, 0.05% Sodium azide and 5% BSA. Preservative 0.05% Sodium azide Stabilizer 5% BSA Concentration 0.5 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/3 Bioinformation Gene Symbol STK11 Gene Full Name serine/threonine kinase 11 Background This gene, which encodes a member of the serine/threonine kinase family, regulates cell polarity and functions as a tumor suppressor. -

Neuronal Migration and Protein Kinases
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Frontiers - Publisher Connector REVIEW ARTICLE published: 13 January 2015 doi: 10.3389/fnins.2014.00458 Neuronal migration and protein kinases Toshio Ohshima* Laboratory for Molecular Brain Science, Department of Life Science and Medical Bioscience, Waseda University, Tokyo, Japan Edited by: The formation of the six-layered structure of the mammalian cortex via the inside-out Kazunori Nakajima, Keio University pattern of neuronal migration is fundamental to neocortical functions. Extracellular cues School of Medicine, Japan such as Reelin induce intracellular signaling cascades through the protein phosphorylation. Reviewed by: Migrating neurons also have intrinsic machineries to regulate cytoskeletal proteins and Mikio Hoshino, National Center of Neurology and Psychiatry, Japan adhesion properties. Protein phosphorylation regulates these processes. Moreover, the Shin-ichi Hisanaga, Tokyo balance between phosphorylation and dephosphorylation is modified by extracellular cues. Metropolitan University, Japan Multipolar-bipolar transition, radial glia-guided locomotion and terminal translocation are *Correspondence: critical steps of radial migration of cortical pyramidal neurons. Protein kinases such as Toshio Ohshima, Laboratory for Cyclin-dependent kinase 5 (Cdk5) and c-Jun N-terminal kinases (JNKs) involve these steps. Molecular Brain Science, Department of Life Science and In this review, I shall give an overview the roles of protein kinases in neuronal migration. Medical Bioscience, Waseda University, 2-2 Wakamatsu-cho, Keywords: protein phosphorylation, kinase, phosphatase, migration, cerebral cortex Shinjuku-ku, Tokyo 162-8480, Japan e-mail: [email protected] CYTOSKELETON DYNAMICS DURING NEURONAL proximal region of the leading process during the locomotion MIGRATION mode of migration (Nishimura et al., 2014). -

Regulation of the Polarity Kinases PAR-1/MARK by 14-3-3 Interaction and Phosphorylation
Research Article 4059 Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation Olga Göransson1,*,‡, Maria Deak1, Stephan Wullschleger1, Nick A. Morrice1, Alan R. Prescott2 and Dario R. Alessi1 1University of Dundee, MRC Protein Phosphorylation Unit, James Black Centre and 2University of Dundee, Division of Cell Biology and Immunology, MSI/WTB complex, Dow Street, Dundee, DD1 5EH, Scotland, UK *Present address: Lund University, BMC, C11, S-22184 Lund, Sweden ‡Author for correspondence (e-mail: [email protected]) Accepted 12 June 2006 Journal of Cell Science 119, 4059-4070 Published by The Company of Biologists 2006 doi:10.1242/jcs.03097 Summary Members of the PAR-1/MARK kinase family play critical membrane. We provide data indicating that the membrane roles in polarity and cell cycle control and are regulated by localisation of MARK3 required a highly conserved C- 14-3-3 scaffolding proteins, as well as the LKB1 tumour terminal domain, which has been termed kinase-associated suppressor kinase and atypical protein kinase C (PKC). domain-1 (KA-1). We also show that dissociation of 14-3-3 In this study, we initially investigated the mechanism from MARK3 did not affect catalytic activity, and that a underlying the interaction of mammalian MARK3 with MARK3 mutant, which could not interact with 14-3-3, was 14-3-3. We demonstrate that 14-3-3 binding to MARK3 normally active. Finally, we establish that there are is dependent on phosphorylation, and necessitates the significant differences in the subcellular localisation of phosphate-binding pocket of 14-3-3. -

A Role for LKB1 Gene in Human Cancer Beyond the Peutz–Jeghers Syndrome
Oncogene (2007) 26, 7825–7832 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc REVIEW A role for LKB1 gene in human cancer beyond the Peutz–Jeghers syndrome M Sanchez-Cespedes Molecular Pathology Programme, Spanish National Cancer Centre (CNIO), Melchor Fernandez Almagro, Madrid, Spain Germline LKB1 mutations are responsible for Peutz– development, LKB1 expression becomes more pro- Jeghers syndrome (PJS). Tumors at several locations nounced in heart, esophagus, pancreas, kidney, colon, frequently arise in these patients, confirming that LKB1 is lung, small intestine and stomach (Luukko et al., 1999; linked to cancer predisposition and is therefore a bona fide Rowan et al., 2000). In adult tissues, levels of LKB1 tumor-suppressor gene. In humans, the LKB1 gene is protein are high in most epithelia, in the follicles and located in the short arm of chromosome 19, which is corpus luteum of the ovary, and the seminiferous tubules frequently deleted in many tumors of sporadic origin. of the testis, in myocytes from skeletal muscle and in glia However, LKB1 alterations in tumors other than those of cells (Rowan et al., 2000; Conde et al., 2007; Figure 1a). PJSare rarely reported. Notably, this is not the case for non-small-cell lung cancer, where nearly half of the tumors harbor somatic and homozygous inactivating mutations in LKB1. The present review considers the LKB1 and Peutz–Jeghers syndrome frequency and pattern of LKB1 gene mutations in LKB1 sporadic cancers of various origins, and the role of the Germline mutations of cause the autosomal encoded protein in cancer development. -
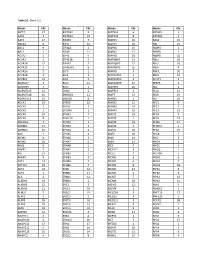
Table S1. Gene List Genes Chr Genes Chr Genes Chr Genes Chr SEPT9
Table S1. Gene List Genes Chr Genes Chr Genes Chr Genes Chr SEPT9 17 EIF2AK3 2 MAPK14 6 RASSF5 1 AAK1 2 EIF2AK4 15 MAPK15 8 RAVER2 1 AATK 17 EIF2B5 3 MAPK3 16 RAX2 19 ABCA1 9 EIF3J 15 MAPK4 18 RB1 13 ABL1 9 EIF4A2 3 MAPK6 15 RBBP4 1 ABL2 1 EIF4B 12 MAPK7 17 RBBP5 1 ACTR2 2 EIF4E 4 MAPK8 10 RBBP8 18 ACVR1 2 EIF4E1B 5 MAPK8IP1 11 RBL1 20 ACVR1B 12 EIF4E2 2 MAPK8IP2 22 RBL2 16 ACVR1C 2 EIF4EBP1 8 MAPK8IP3 16 RBPJ 4 ACVR2A 2 ELF3 1 MAPK9 5 RBPJL 20 ACVR2B 3 ELF4 X MAPKAPK2 1 RBX1 22 ACVRL1 12 ELK1 X MAPKAPK3 3 RCHY1 4 ADAM17 2 ELK3 12 MAPKAPK5 12 REEP5 5 ADAM29 4 ELK4 1 MAPRE1 20 REL 2 ADAMTS15 11 EML4 2 MAPRE3 2 RELA 11 ADAMTS18 16 ENDOD1 11 MAPT 17 RELB 19 ADAMTSL3 15 ENDOG 9 MARK1 1 RET 10 ADCK1 14 EP300 22 MARK2 11 RFC1 4 ADCK2 7 EPAS1 2 MARK3 14 RFC2 7 ADCK3 1 EPCAM 2 MARK4 19 RFC3 13 ADCK4 19 EPHA1 7 MAST1 19 RFC4 3 ADCK5 8 EPHA10 1 MAST2 1 RFC5 12 ADORA1 1 EPHA2 1 MAST3 19 RFNG 17 ADRBK1 11 EPHA3 3 MAST4 5 RFWD2 1 ADRBK2 22 EPHA4 2 MASTL 10 RFX2 19 AGK 7 EPHA5 4 MATK 19 RHEB 7 AIFM1 X EPHA6 3 MAX 14 RHO 3 AIFM2 10 EPHA7 6 MBIP 14 RHOA 3 AIM1 6 EPHA8 1 MCC 5 RHOC 1 AIMP2 7 EPHB1 3 MCF2L2 3 RHOQ 2 AIP 11 EPHB2 1 MCL1 1 RICTOR 5 AKAP4 X EPHB3 3 MCM2 3 RIOK1 6 AKT1 14 EPHB4 7 MCM3 6 RIOK2 5 AKT1S1 19 EPHB6 7 MCM4 8 RIOK3 18 AKT2 19 EPS8 12 MCM5 22 RIPK1 6 AKT3 1 ERBB2 17 MCM6 2 RIPK2 8 ALK 2 ERBB3 12 MCM7 7 RIPK3 14 ALKBH1 14 ERBB4 2 MCM8 20 RIPK4 21 ALKBH2 12 ERC2 3 MDM2 12 RMI1 9 ALKBH3 11 ERCC1 19 MDM4 1 RNASEL 1 ALMS1 2 ERCC2 19 MECOM 3 RNF213 17 ALPK1 4 ERCC3 2 MED12 X RNF220 1 ALPK2 18 ERCC4 16 MED12L 3 ROCK1 18 ALPK3 15 ERCC5 13 -

Bi-Phospho-GSK3B(S21/29) Antibody Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap3111a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Bi-Phospho-GSK3B(S21/29) Antibody Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP3111a Specification Bi-Phospho-GSK3B(S21/29) Antibody - Product Information Application WB, IHC-P,E Primary Accession P49841 Reactivity Human, Mouse Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Bi-Phospho-GSK3B(S21/29) Antibody - Additional Information Gene ID 2932 Other Names Glycogen synthase kinase-3 beta, GSK-3 The anti-Phospho-GSK3B-S21/29 Pab (Cat. beta, Serine/threonine-protein kinase #AP3111a) is used in Western blot to detect GSK3B, GSK3B Phospho-GSK3B-S21/29 in mouse thymus tissue lysate. Target/Specificity This GSK3B Antibody is generated from rabbits immunized with a KLH conjugated synthetic phosphopeptide corresponding to amino acid residues surrounding S21/29 of human GSK3B. Dilution WB~~1:1000 IHC-P~~1:50~100 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Formalin-fixed and paraffin-embedded human cancer tissue reacted with the Storage primary antibody, which was Maintain refrigerated at 2-8°C for up to 2 peroxidase-conjugated to the secondary weeks. For long term storage store at -20°C antibody, followed by AEC staining. This data in small aliquots to prevent freeze-thaw demonstrates the use of this antibody for cycles. immunohistochemistry; clinical relevance has not been evaluated. BC = breast carcinoma; Precautions HC = hepatocarcinoma. Bi-Phospho-GSK3B(S21/29) Antibody is for research use only and not for use in Page 1/4 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 diagnostic or therapeutic procedures. -

On the Road from Gene to Therapy in Inherited Cardiomyopathies
On the road from gene to therapy in Inherited Cardiomyopathies Limongelli Giuseppe1,2,3, Bossone Edoardo4, Elliott Perry Mark2,3, Day Sharlene5 1 Department of Cardiothoracic Sciences, Università della Campania “Luigi Vanvitelli”, Monaldi Hospital, AORN Colli, Naples, Italy 2 Institute of Cardiovascular Science, University College of London, London, UK 3 European Reference Network - GUARD HEART 4 Cardiology Division, University of Salerno, Salerno, Italy 5 Departments of Internal Medicine and Pediatrics, University of Michigan, Ann Arbor, Michigan, USA Key Words: inherited cardiomyopathies, genetics, therapy, precision medicine Summary: Almost 30 years after the discovery of the first genetic mutation for HCM, the deve- lopment of new pharmacological approaches targeting cardiomyopathies and other orphan/rare car- diac disease is becoming closer to reality. Development of targeted therapies is enabled by new in- sights into the clinical and molecular aspects, and pathogenesis of cardiomyopathies, along with the establishment of large-scale international collaboration and the increasing engagement of the phar- maceutical industry. The road toward “cardiovascular precision medicine” is just beginning, with inherited and rare diseases leading the way in this exciting new era. Key Points (3–5) • Cardiomyopathies are a heterogeneous group of myocardial disorders • Most are related to genetic abnormalities of the structural and functional proteins of the myocyte • The clinical course recognize 3 phases: preclinical, overt (concealed), and end stage disease • New insight on pathogenesis and mechanisms underlying inherited cardiomyopathies is opening the hope of present and future therapies Address for correspondence: Giuseppe Limongelli, MD, PhD, FESC Department of Cardiothoracic Sciences, Università della Campania “Luigi Vanvitelli”, Monaldi Hospital, AORN Colli, Naples, Italy Email: [email protected] The century that closed the second millennium was marked by enormous progress in all the life sci- ences, including medicine. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

Gene Expression Profile Associated with Response to Doxorubicin
Cancer Therapy: Clinical Gene Expression Profile Associated with Response to Doxorubicin-Based Therapy in Breast Cancer Maria Aparecida Azevedo Koike Folgueira,1Dirce Maria Carraro,4 Helena Brentani,4 Diogo Ferreira da Costa Patra‹ o,5 Edson Mantovani Barbosa,6 Ma¤ rio Moura‹ oNetto,5 Jose¤ Roberto Fı´garo Caldeira,8 Maria Lucia Hirata Katayama,1Fernando Augusto Soares,5 Ce¤ lia Tosello Oliveira,6 Luiz Fernando Lima Reis,4 Jane Haruko Lima Kaiano,4 Luiz Paulo Camargo,4 Ricardo Zorzetto NicolielloVe“ ncio,7 Igor Moyse¤ s Longo Snitcovsky,2 Fabiana Baroni Alves Makdissi,5 Paulo Jose¤ da Silva e Silva,3 Joa‹ o Carlos Guedes Sampaio Go¤ es,6 and Maria Mitzi Brentani1 Abstract Purpose:This study was designed to identify genes that could predict response to doxorubicin- based primary chemotherapy in breast cancer patients. Experimental Design: Biopsy samples were obtained before primary treatment with doxorubi- cin and cyclophosphamide. RNA was extracted and amplified and gene expression was analyzed using cDNA microarrays. Results: Response to chemotherapy was evaluated in 51patients, and based on Response Eval- uation Criteria in Solid Tumors guidelines, 42 patients, who presented at least a partial response (z30% reduction in tumor dimension), were classified as responsive. Gene profile of samples, divided into training set (n = 38) and independent validation set (n = 13), were at first analyzed against a cDNA microarray platform containing 692 genes. Unsupervised clustering could not separate responders from nonresponders. A classifier was identified comprising EMILIN1, FAM14B,andPBEF, which however could not correctly classify samples included in the validation set. Our next step was to analyze gene profile in a more comprehensive cDNA microarray platform, containing 4,608 open reading frame expressed sequence tags.