Using Parallel Execution Perl Program on Multiple Biohpc Lab Machines
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CIS 90 - Lesson 2
CIS 90 - Lesson 2 Lesson Module Status • Slides - draft • Properties - done • Flash cards - NA • First minute quiz - done • Web calendar summary - done • Web book pages - gillay done • Commands - done • Lab tested – done • Print latest class roster - na • Opus accounts created for students submitting Lab 1 - • CCC Confer room whiteboard – done • Check that headset is charged - done • Backup headset charged - done • Backup slides, CCC info, handouts on flash drive - done 1 CIS 90 - Lesson 2 [ ] Has the phone bridge been added? [ ] Is recording on? [ ] Does the phone bridge have the mike? [ ] Share slides, putty, VB, eko and Chrome [ ] Disable spelling on PowerPoint 2 CIS 90 - Lesson 2 Instructor: Rich Simms Dial-in: 888-450-4821 Passcode: 761867 Emanuel Tanner Merrick Quinton Christopher Zachary Bobby Craig Jeff Yu-Chen Greg L Tommy Eric Dan M Geoffrey Marisol Jason P David Josh ? ? ? ? Leobardo Gabriel Jesse Tajvia Daniel W Jason W Terry? James? Glenn? Aroshani? ? ? ? ? ? ? = need to add (with add code) to enroll in Ken? Luis? Arturo? Greg M? Ian? this course Email me ([email protected]) a relatively current photo of your face for 3 points extra credit CIS 90 - Lesson 2 First Minute Quiz Please close your books, notes, lesson materials, forum and answer these questions in the order shown: 1. What command shows the other users logged in to the computer? 2. What is the lowest level, inner-most component of a UNIX/Linux Operating System called? 3. What part of UNIX/Linux is both a user interface and a programming language? email answers to: [email protected] 4 CIS 90 - Lesson 2 Commands Objectives Agenda • Understand how the UNIX login • Quiz operation works. -
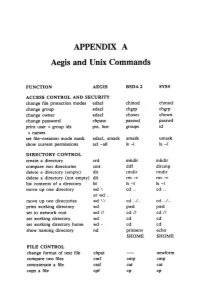
APPENDIX a Aegis and Unix Commands
APPENDIX A Aegis and Unix Commands FUNCTION AEGIS BSD4.2 SYSS ACCESS CONTROL AND SECURITY change file protection modes edacl chmod chmod change group edacl chgrp chgrp change owner edacl chown chown change password chpass passwd passwd print user + group ids pst, lusr groups id +names set file-creation mode mask edacl, umask umask umask show current permissions acl -all Is -I Is -I DIRECTORY CONTROL create a directory crd mkdir mkdir compare two directories cmt diff dircmp delete a directory (empty) dlt rmdir rmdir delete a directory (not empty) dlt rm -r rm -r list contents of a directory ld Is -I Is -I move up one directory wd \ cd .. cd .. or wd .. move up two directories wd \\ cd . ./ .. cd . ./ .. print working directory wd pwd pwd set to network root wd II cd II cd II set working directory wd cd cd set working directory home wd- cd cd show naming directory nd printenv echo $HOME $HOME FILE CONTROL change format of text file chpat newform compare two files emf cmp cmp concatenate a file catf cat cat copy a file cpf cp cp Using and Administering an Apollo Network 265 copy std input to std output tee tee tee + files create a (symbolic) link crl In -s In -s delete a file dlf rm rm maintain an archive a ref ar ar move a file mvf mv mv dump a file dmpf od od print checksum and block- salvol -a sum sum -count of file rename a file chn mv mv search a file for a pattern fpat grep grep search or reject lines cmsrf comm comm common to 2 sorted files translate characters tic tr tr SHELL SCRIPT TOOLS condition evaluation tools existf test test -
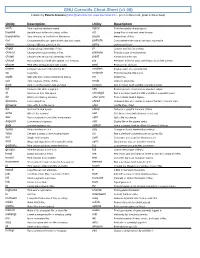
GNU Coreutils Cheat Sheet (V1.00) Created by Peteris Krumins ([email protected], -- Good Coders Code, Great Coders Reuse)
GNU Coreutils Cheat Sheet (v1.00) Created by Peteris Krumins ([email protected], www.catonmat.net -- good coders code, great coders reuse) Utility Description Utility Description arch Print machine hardware name nproc Print the number of processors base64 Base64 encode/decode strings or files od Dump files in octal and other formats basename Strip directory and suffix from file names paste Merge lines of files cat Concatenate files and print on the standard output pathchk Check whether file names are valid or portable chcon Change SELinux context of file pinky Lightweight finger chgrp Change group ownership of files pr Convert text files for printing chmod Change permission modes of files printenv Print all or part of environment chown Change user and group ownership of files printf Format and print data chroot Run command or shell with special root directory ptx Permuted index for GNU, with keywords in their context cksum Print CRC checksum and byte counts pwd Print current directory comm Compare two sorted files line by line readlink Display value of a symbolic link cp Copy files realpath Print the resolved file name csplit Split a file into context-determined pieces rm Delete files cut Remove parts of lines of files rmdir Remove directories date Print or set the system date and time runcon Run command with specified security context dd Convert a file while copying it seq Print sequence of numbers to standard output df Summarize free disk space setuidgid Run a command with the UID and GID of a specified user dir Briefly list directory -

Lab 2: Using Commands
Lab 2: Using Commands The purpose of this lab is to explore command usage with the shell and miscellaneous UNIX commands. Preparation Everything you need to do this lab can be found in the Lesson 2 materials on the CIS 90 Calendar: http://simms-teach.com/cis90calendar.php. Review carefully all Lesson 2 slides, even those that may not have been covered in class. Check the forum at: http://oslab.cis.cabrillo.edu/forum/ for any tips and updates related to this lab. The forum is also a good place to ask questions if you get stuck or help others. If you would like some additional assistance come to the CIS Lab on campus where you can get help from instructors and student lab assistants: http://webhawks.org/~cislab/. Procedure Please log into the Opus server at oslab.cis.cabrillo.edu via port 2220. You will need to use the following commands in this lab. banner clear finger man uname bash date history passwd whatis bc echo id ps who cal exit info type Only your command history along with the three answers asked for by the submit script will be graded. You must issue each command below (exactly). Rather than submitting answers to any questions asked below you must instead issue the correct commands to answer them. Your command history will be scanned to verify each step was completed. The Shell 1. What shell are you currently using? What command did you use to determine this? (Hint: We did this in Lab 1) 2. The type command shows where a command is located. -

Scripting in Axis Network Cameras and Video Servers
Scripting in Axis Network Cameras and Video Servers Table of Contents 1 INTRODUCTION .............................................................................................................5 2 EMBEDDED SCRIPTS ....................................................................................................6 2.1 PHP .....................................................................................................................................6 2.2 SHELL ..................................................................................................................................7 3 USING SCRIPTS IN AXIS CAMERA/VIDEO PRODUCTS ......................................8 3.1 UPLOADING SCRIPTS TO THE CAMERA/VIDEO SERVER:...................................................8 3.2 RUNNING SCRIPTS WITH THE TASK SCHEDULER...............................................................8 3.2.1 Syntax for /etc/task.list.....................................................................................................9 3.3 RUNNING SCRIPTS VIA A WEB SERVER..............................................................................11 3.3.1 To enable Telnet support ...............................................................................................12 3.4 INCLUDED HELPER APPLICATIONS ..................................................................................13 3.4.1 The image buffer - bufferd........................................................................................13 3.4.2 sftpclient.........................................................................................................................16 -

CIS 192 Linux Lab Exercise
CIS 192 Linux Lab Exercise Lab 2: Joining a network Spring 2013 Lab 2: Joining a network The purpose of this lab is to configure permanently the network settings of several systems to join one or more networks. This includes setting the IP address, network mask, default gateway, and DNS settings for different distributions of Linux. Once joined, the connectivity will be tested and network traffic observed. Supplies Frodo, Elrond and William VMs Forum Use the forum to ask for help, post tips and any lessons learned when you have finished. Forum is at: http://oslab.cabrillo.edu/forum/ Background For a Linux system to join a LAN (Local Area Network) an IP address and subnet mask must be configured on one of its NIC interfaces. The IP address can be dynamic (obtained automatically from a DHCP server) or static (manually configured IP address). To reach beyond the LAN to the Internet, a default gateway must be configured. To be able to use hostnames, e.g. google.com, rather than numerical IP address a DNS name server must be configured. The IP and gateway settings can be configured temporarily with the ifconfig and route commands. These settings will remain in place till the system or network service is restarted. For a permanent solution, the IP and gateway information must be added to the appropriate network configuration files in the /etc directory. These files are used by the network service during system startup. Finally, there are a number of commands (utilities) that can be used to check that traffic is flowing correctly on the networks. -

PHYS 210: Introduction to Computational Physics Fall 2014 Homework 1: Version 6—September 23, 2014—Problem 2, Item 6 Description Amended
PHYS 210: Introduction to Computational Physics Fall 2014 Homework 1: Version 6—September 23, 2014—Problem 2, item 6 description amended. Due: Thursday, September 25, 11:59 PM PLEASE report all bug reports, comments, gripes etc. to Matt: [email protected] Please make careful note of the following information and instructions, much of which will also apply to subsequent assignments. 1. There are 5 problems in this homework, most of which have multiple parts. 2. Problems marked with (⋆) may be particularly challenging, so don’t be surprised if it takes you longer to figure out solutions for them than the others. 3. Please do not be put off / terrified etc. by the length of the handout, including this preamble. As previous students of my computational physics courses can attest, I tend to spell things out in gory detail, so there really isn’t as much work to do as it might seem. 4. As discussed in the lab, I have created directories for all of you of the form /phys210/$LOGNAME, where $LOGNAME is the name of your PHAS account. 5. Within /phys210/$LOGNAME, I have also created sub-directories hw1, hw2 and hw3, which you will use to complete the three homework assignments in this course. In particular, for this assignment, you will create various directories and files that will need to reside within /phys210/$LOGNAME/hw1, and any reference to directory hw1 below is implicitly a reference to the absolute pathname /phys210/$LOGNAME/hw1. 6. The hw[1-3] sub-directories are read, write and execute protected from other users. -

USER MANUAL GWR High Speed Cellular Router Series
GWR USER MANUAL GWR High Speed Cellular Router Series Document version 1.0.0 Date: December 2015 WWW.GENEKO.RS User Manual Document History Date Description Author Comments 24.12.2015 User Manual Tanja Savić Firmware versions: 1.1.2 Document Approval The following report has been accepted and approved by the following: Signature Printed Name Title Date Dragan Marković Executive Director 24.12.2015 GWR High Speed Cellular Router Series 2 User Manual Content DOCUMENT APPROVAL ........................................................................................................................................ 2 LIST OF FIGURES .................................................................................................................................................... 5 LIST OF TABLES ...................................................................................................................................................... 8 DESCRIPTION OF THE GPRS/EDGE/HSPA ROUTER SERIES ...................................................................... 9 TYPICAL APPLICATION ............................................................................................................................... 10 TECHNICAL PARAMETERS ......................................................................................................................... 11 PROTOCOLS AND FEATURES ...................................................................................................................... 14 PRODUCT OVERVIEW ............................................................................................................................... -

Gnu Coreutils Core GNU Utilities for Version 5.93, 2 November 2005
gnu Coreutils Core GNU utilities for version 5.93, 2 November 2005 David MacKenzie et al. This manual documents version 5.93 of the gnu core utilities, including the standard pro- grams for text and file manipulation. Copyright c 1994, 1995, 1996, 2000, 2001, 2002, 2003, 2004, 2005 Free Software Foundation, Inc. Permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.1 or any later version published by the Free Software Foundation; with no Invariant Sections, with no Front-Cover Texts, and with no Back-Cover Texts. A copy of the license is included in the section entitled “GNU Free Documentation License”. Chapter 1: Introduction 1 1 Introduction This manual is a work in progress: many sections make no attempt to explain basic concepts in a way suitable for novices. Thus, if you are interested, please get involved in improving this manual. The entire gnu community will benefit. The gnu utilities documented here are mostly compatible with the POSIX standard. Please report bugs to [email protected]. Remember to include the version number, machine architecture, input files, and any other information needed to reproduce the bug: your input, what you expected, what you got, and why it is wrong. Diffs are welcome, but please include a description of the problem as well, since this is sometimes difficult to infer. See section “Bugs” in Using and Porting GNU CC. This manual was originally derived from the Unix man pages in the distributions, which were written by David MacKenzie and updated by Jim Meyering. -

1. UNIX-Tools
UNIX Tools Akadia AG 1. UNIX-Tools In diesem abschliessenden Kapitel sollen einige Anregungen aus der Praxis vermittelt werden. Der Kurs wird schliesslich mit der Betrachtung einiger UNIX-Tools wie grep, sed, tr, find und der Vorstellung einiger Public Domain Tools abgerundet. Ein kurzer Aus- blick in die UNIX-Zukunft soll verdeutlichen, dass UNIX kein «totes» Betriebssystem ist, sondern von verschiedenen Firmen, Gremien, Universitäten und Konsortien weiterent- wickelt wird. 1.1 Oft benutzte Utilities 1.1.1 find (find files by name, or by other characteristics) Das Kommando find(1) ist eines der unter UNIX am häufigsten benutzten Utilities. Es durchsucht das Filesystem, ausgehend von einem Startpunkt, rekursiv nach Filenamen, welche nach bestimmten Kriterien ausgewählt werden. Die häufigsten Anwendungen von find sind: • Generieren einer Liste von Files zur Datensicherung • Durchforsten des Filesystems nach einem «verlorenen» File • Durchsuchen des Fileinhalts nach einem bestimmten Wort oder Pattern Directories in Filesystem auflisten $ find . -type d -print Alle Files mit der Endung (.o) in Filesystem löschen $ find . -name "*.o" -exec rm -i {} \; Suchen aller Files die jünger sind als ein Vergleichsfile $ find . -newer <vergleichsfile> -print Löschen aller "core" und "*.BAK" Files $ find . (-name core -o -name ’*.BAK’) -exec rm -f {} \; Suchen aller Files die in den letzten 7 Tagen verändert wurden $ find . ! -mtime +7 -print Suchen aller Files die in den letzten 7 Tagen NICHT verändert wurden $ find . -mtime +7 -print Suchen aller Files welche "pattern" enthalten $ find . -type f -exec grep -l "stdio.h" {} \; ./Xm/examples/dogs/Dog.c ./Xm/examples/dogs/Square.c ......... Suchen aller Files welche "pattern" enthalten mit Angabe der Zeile $ find . -

Chapter 1 Introducing UNIX
Chapter 9 The Shell – Customizing the Environment Tien-Hsiung Weng 翁添雄 [email protected] Objectives • Know the difference between local and environment variables • Examine PATH, SHELL, MAIL, etc • Use the history mechanism to recall, edit, and run previously execute commands • Prevent accidental overwriting of files and logging out using set –o The Shell • The Unix shell is both an interpreter and a scripting language • When log in, an interactive shell presents a prompt and waits for requests • Shell supports job control, aliases, and history • An interactive shell runs a non-interactive shell when executing a shell script • C shell was created by Billy Joy • To know the shell we are using: echo $SHELL • We can run chsh command to change the entry in /etc/passwd (non-linux) • Make a temporary switch by running the shell itself as a command: csh (C shell runs as a child) and exit (terminate C shell back to login shell) Environment variables env command displays only environment variables PATH, SHELL, HOME, LOGNAME, USER, and so on, are environment variables MY_DIR=/home/eric/temp echo $MY_DIR sh echo $MY_DIR set will display the value of MY_DIR, but not env export MY_DIR (in Bourne and BASH) export statement enforces variable inheritance setenv MY_DIR (in C shell to enforce variable inheritance) Common Environment Variables HOME Æ Home dir (the directory a user is placed on logging in) PATH Æ List of directories searched by shell to locate a command LOGNAME Æ Login name of user USER Æ as above MAIL Æ Absolute pathname of user’s mailbox file MAILCHECK Æ Mail checking interval for incoming mail TERM Æ Type of terminal PWD Æ Absolute pathname of current directory (Korn and BASH only) CDPATH Æ List of directories searched by cd when used with a non-absolute pathname PS1 Æ Primary prompt string PS2 Æ Secondary prompt string SHELL Æ User’s login shell and one invoked by programs having shell escapes Prompt strings (PS1, PS2, PWD) echo $PS1 PS1=“C>” (in BASH) C> PS1=‘[$PWD]’ PS1=“\h>” (\h Æ hostname) Normally, PS2 is set to > or $ find . -

Lab Work 05. Linux/UNIX Shell Environment Variables
LAB WORK 05. LINUX/UNIX SHELL ENVIRONMENT VARIABLES 1. PURPOSE OF WORK • Learn to use environment variables and assign new values to them; • Consolidate skills in the use of elementary commands and utilities of UNIX/Linux; • Get the concept of initialization program files using the shell example; • Acquire skills in creating and debugging shell configuration files. The Linux/UNIX commands are used: sudo, adduser, userdel, set, unset, readonly, export, env, more, less, man, ls, echo, pwd, wc, who, date, ... 2. TASKS FOR WORK NOTE. Start Your UbuntuMini Virtual Machine on your VirtualBox. You need only Linux Terminal to complete the lab tasks. Before completing the tasks, make a SnapShot of your Virtual Linux. If there are problems, you can easily go back to working condition! 2.1. Shell Environment Variables Analyze. (Fill in a Table) 2.2. Make Example bash config file. (Make a config-file Screenshot) 2.3. Generate Your Task Variant Nr. (Make a Task Text Copy) 2.4. Change 3 Shell Config Files (according to your variant). (Make 3 Text Copy of config-files) © Yuriy Shamshin, 2020 1/17 3. REPORT Make a report about this work and send it to the teacher’s email (use a docx Report Blank). REPORT FOR LAB WORK 05: LINUX/UNIX SHELL ENVIRONMENT VARIABLES Student Name Surname Student ID (nV) Date 3.1. Insert Completing Shell Environment Variables Analyze Table, 3.2. Insert Screenshot with Your Config-file and working Clock, 3.3. Insert Text Copy of Task for Your Variant, 3.4. Insert Text Copy of Your Modification for 3 Configuration Files with the necessary comments: ˜/.bash_profile, ˜/.bashrc, ˜/.bash_logout.