Effect of Geographic Range Discontinuity on Species Differentiation—East-Mediterranean Abies Cilicica:Acasestudy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ISTA List of Stabilized Plant Names 7Th Edition
ISTA List of Stabilized Plant Names th 7 Edition ISTA Nomenclature Committee Chair: Dr. M. Schori Published by All rights reserved. No part of this publication may be The Internation Seed Testing Association (ISTA) reproduced, stored in any retrieval system or transmitted Zürichstr. 50, CH-8303 Bassersdorf, Switzerland in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior ©2020 International Seed Testing Association (ISTA) permission in writing from ISTA. ISBN 978-3-906549-77-4 ISTA List of Stabilized Plant Names 1st Edition 1966 ISTA Nomenclature Committee Chair: Prof P. A. Linehan 2nd Edition 1983 ISTA Nomenclature Committee Chair: Dr. H. Pirson 3rd Edition 1988 ISTA Nomenclature Committee Chair: Dr. W. A. Brandenburg 4th Edition 2001 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 5th Edition 2007 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 6th Edition 2013 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 7th Edition 2019 ISTA Nomenclature Committee Chair: Dr. M. Schori 2 7th Edition ISTA List of Stabilized Plant Names Content Preface .......................................................................................................................................................... 4 Acknowledgements ....................................................................................................................................... 6 Symbols and Abbreviations .......................................................................................................................... -

Genetics and Evolution of the Mediterranean Abies Species
Acta Universitatis Agriculturae sueciae SlLVESTRIA 148 > z SLU Genetics and Evolution of the Mediterranean Abies Species Laura Parducci Genetics and Evolution of the Mediterranean Abies species Laura Parducci Akademiska avhandling som för vinnande av filosofie doktorsexamen kommer att offentlig försvaras i hörsal Björken, SLU, fredagen den 8 september 2000, kl. 10.00. Abstract This thesis summarizes and discusses results o f five separate studies in which molecular techniques have been used to study the genetic variability and evolution o f the Abies taxa occurring in the Mediterranean region. In particular, the investigation focused on the rare speciesAbies nebrodensis (Lojac.) Mattei, endemic to the island of Sicily, and the three neighbouring speciesA. alba (Mill.), A. cephalonica (Laud.) and A. numidica (De Lann.). The main aim o f the studies was to determine the amount and distribution of the genetic variability within and among Mediterranean taxa ofAbies, at both the nuclear and chloroplast levels, in order to elucidate their origin and evolution and to shed light on the taxonomic position o f A. nebrodensis. In studies I, II and V allozyme markers were used to provide information on the level and distribution o f genetic variation among and within natural populations ofA. alba, A. cephalonica, A. nebrodensis and A. numidica and to estimate the outcrossing rate within A. alba. In studies III and IV, DNA markers from the chloroplast genome were developed and employed at the intra- and inter-specific levels to estimate the degree of cpDNA variation in the genus and to derive inferences concerning species relationships. Two different approaches were used: the first involved a comparative restriction-site analysis of ten different amplified chloroplast DNA fragments and the second involved the analysis o f six chloroplast hypervariable repetitive simple-sequence repeats (cpSSRs or microsatellites). -

Effect of Geographic Range Discontinuity on Taxonomic Differentiation of Abies Cilicica
Acta Societatis Botanicorum Poloniae ORIGINAL RESEARCH PAPER Acta Soc Bot Pol 84(4):419–430 DOI: 10.5586/asbp.2015.037 Received: 2015-05-26 Accepted: 2015-10-29 Published electronically: 2015-12-31 Effect of geographic range discontinuity on taxonomic differentiation of Abies cilicica Krystyna Boratyńska1, Katarzyna Sękiewicz1, Anna Katarzyna Jasińska1, Dominik Tomaszewski1, Grzegorz Iszkuło1,2, Tolga Ok3, Magda Bou Dagher-Kharrat4, Adam Boratyński1* 1 Institute of Dendrology of the Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland 2 University of Zielona Góra, Faculty of Biological Sciences, Prof. Z. Szafrana 1, 65-516 Zielona Góra, Poland 3 Department of Forest Botany, Faculty of Forestry, Kahramanmaras Sutcu Imam University, 46100 Kahramanmaras, Turkey 4 Laboratoire ‘Caractérisation Génomique des Plantes’, Faculté des Sciences, Université Saint-Joseph, Campus Sciences et Technologies, Mar Roukos, Mkalles, BP: 1514 Riad el Solh, Beirut 1107 2050, Lebanon Abstract Three populations of Abies cilicica subsp. isaurica and four of A. cilicica subsp. cilicica were analyzed using 35 morphologi- cal and anatomical needle characters with the implementation of multivariate statistical methods to verify the differences between subspecies. Moreover, the possible geographic differentiation ofA. cilicica subsp. cilicica populations from the East Taurus and Lebanon Mountains was examined. Abies cilicica subsp. isaurica has been distinguished from A. cilicica subsp. cilicica by its glabrous young shoots and resinous buds. We detected that needles of A. cilicica subsp. isaurica are longer, broader and thicker, with a higher number of stomata rows, and larger cells of the epidermis, hypodermis and endodermis than A. cilicica subsp. cilicica. Additionally, A. cilicica subsp. isaurica needles have frequently rounded to obtuse-acute apex and resinous canals positioned more centrally inside the mesophyll than needles of A. -

The Evolution of Cavitation Resistance in Conifers Maximilian Larter
The evolution of cavitation resistance in conifers Maximilian Larter To cite this version: Maximilian Larter. The evolution of cavitation resistance in conifers. Bioclimatology. Univer- sit´ede Bordeaux, 2016. English. <NNT : 2016BORD0103>. <tel-01375936> HAL Id: tel-01375936 https://tel.archives-ouvertes.fr/tel-01375936 Submitted on 3 Oct 2016 HAL is a multi-disciplinary open access L'archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destin´eeau d´ep^otet `ala diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publi´esou non, lished or not. The documents may come from ´emanant des ´etablissements d'enseignement et de teaching and research institutions in France or recherche fran¸caisou ´etrangers,des laboratoires abroad, or from public or private research centers. publics ou priv´es. THESE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITE DE BORDEAUX Spécialité : Ecologie évolutive, fonctionnelle et des communautés Ecole doctorale: Sciences et Environnements Evolution de la résistance à la cavitation chez les conifères The evolution of cavitation resistance in conifers Maximilian LARTER Directeur : Sylvain DELZON (DR INRA) Co-Directeur : Jean-Christophe DOMEC (Professeur, BSA) Soutenue le 22/07/2016 Devant le jury composé de : Rapporteurs : Mme Amy ZANNE, Prof., George Washington University Mr Jordi MARTINEZ VILALTA, Prof., Universitat Autonoma de Barcelona Examinateurs : Mme Lisa WINGATE, CR INRA, UMR ISPA, Bordeaux Mr Jérôme CHAVE, DR CNRS, UMR EDB, Toulouse i ii Abstract Title: The evolution of cavitation resistance in conifers Abstract Forests worldwide are at increased risk of widespread mortality due to intense drought under current and future climate change. -

Antibacterial Activity of Spruce Bark (Picea Abies L.) Extract Against Escherichia Coli
ISSN: 2601 – 6141, ISSN-L: 2601 – 6141 Acta Biologica Marisiensis 2018, 1(1): 5-9 ORIGINAL PAPER ANTIBACTERIAL ACTIVITY OF SPRUCE BARK (PICEA ABIES L.) EXTRACT AGAINST ESCHERICHIA COLI Corneliu TANASE1, Irina BOZ2,3*, Silvia OROIAN1, Sanda COȘARCĂ1, Felicia TOMA1, Anca MARE1, Adrian MAN1 1University of Medicine and Pharmacy of Tîrgu Mureș, Gheorghe Marinescu, 38, 540139, Tîrgu Mureș, Mureș, Romania 2NIRDBS - Institute of Biological Research, Department of Experimental and applied biology, Lascăr Catargi 47,700107, Iași, Romania 3Integrated Centre for Environmental Science Studies in the North-East Development Region – CERNESIM, Alexandru Ioan Cuza University, Carol I 20 A, 700505 Iași, Romania *Correspondence: Irina BOZ [email protected] Received: 17 May 2018; Accepted: 17 June 2018; Published: 15 July 2018 Abstract: The increase of antibiotic resistant bacteria in lasts years resulted in limited options for treatment of bacterial diseases. Escherichia coli is one of the most common Gram-negative bacterial pathogen and a cause of both community and hospital acquired infections. Medicinal plants are alternative rich sources of useful antibacterial agents. The antimicrobial activities of the spruce (Picea abies L.) bark polyphenolic extracts were examined for their capacity to inhibit the growth of Escherichia coli. Spruce bark extract was obtained by conventional aqueous extraction and with ultrasounds. The minimum inhibitory concentration was determined by microdilution method. The antibacterial effect of both extracts was strong against Escherichia coli. The antimicrobial effect of polyphenolic extracts on Escherichia coli was expressed at a concentration of 15 mg/ml. Antimicrobial activity of spruce bark compounds suggest a possible use of spruce bark in pharmaceutical preparations. -

Cedrus Libani A
Güney et al. Botanical Studies (2015) 56:20 DOI 10.1186/s40529-015-0100-z RESEARCH Open Access Cambial activity and xylogenesis in stems of Cedrus libani A. Rich at different altitudes Aylin Güney*, Danielle Kerr, Ayça Sökücü, Reiner Zimmermann and Manfred Küppers Abstract Background: The dynamics of cambial activity and xylogenesis provide information on how and to what extent wood formation respond to climatic variability. The Lebanon Cedar (Cedrus libani A.Rich) is a montane tree species which is distributed along a wide altitudinal range in the northeastern Mediterranean region, currently considered as a potential forest species for Central Europe with respect to climate change. This study provides first data on intra-annual growth dynamics at cellular level using the microcore technique for a montane Mediterranean tree species at different altitudes within and outside its natural range. Results: Microcores were collected fortnightly in the growing season of 2013 in order to study temporal dynamics of cambial activity and xylogenesis in stems of C. libani at different altitudes in the Taurus Mountains (1000 – 2000 m a.s.l.) and at a plantation at Bayreuth (330 m a.s.l.; Germany). The dormant cambium consisted of about 5 cells at the Turkish sites and 7 cells at Bayreuth. Cambial activity set in, when daily minimum temperatures exceeded 0 °C and daily means of air and stem temperature exceeded 5 °C. Xylogenesis started between April and May, ended approximately the end of September to the beginning of October and lasted 134 (at tree line) to 174 days (at the lowest Turkish site). -

Wood Anatomy of the Genus Abies Luis García Esteban*, Paloma
IAWA Journal, Vol. 30 (3), 2009: 231–245 WOOD ANATOMY OF THE GENUS ABIES A REVIEW Luis García Esteban*, Paloma de Palacios, Francisco García Fernández and Ruth Moreno Universidad Politécnica de Madrid. Escuela Técnica Superior de Ingenieros de Montes, Departamento de Ingeniería Forestal, Ciudad Universitaria, 28040 Madrid, Spain *Corresponding author [E-mail: [email protected]] SUMMARY The literature on the wood anatomy of the genus Abies is reviewed and discussed, and complemented with a detailed study of 33 species, 1 sub- species and 4 varieties. In general, the species studied do not show diag- nostic interspecific differences, although it is possible to establish differences between groups of species using certain quantitative and quali- tative features. The marginal axial parenchyma consisting of single cells and the ray parenchyma cells with distinctly pitted horizontal walls, nodular end walls and presence of indentures are constant for the genus, although these features also occur in the other genera of the Abietoideae. The absence of ray tracheids in Abies can be used to distinguish it from Cedrus and Tsuga, and the irregularly shaped parenchymatous marginal ray cells are only shared with Cedrus. The absence of resin canals enables Abies to be distinguished from very closely related genera such as Keteleeria and Nothotsuga. The crystals in the ray cells, taxodioid cross-field pitting and the warty layer in the tracheids can be regarded as diagnostic generic features. Key words: Abies, Abietoideae, anatomy, wood. INTRODUCTION The family Pinaceae, with 11 genera and 225 species, is the largest conifer family. The genus Abies, with 48 species and 24 varieties, has the second highest number of species after the genus Pinus (Farjon 2001). -

Spruces, Firs and Larches 1449 Established Using Genetically Modified P
TEMPERATE ECOSYSTEMS / Spruces, Firs and Larches 1449 established using genetically modified P. tremula, Spruces, Firs and Larches P. tremuloides, P. deltoides,andP. alba  P. tremula selected varieties. The use of genetically modified H Wolf, State Board for Forestry (Saxony), Pirna, plants has raised concerns over the risk posed to Germany the fitness or future adaptability of wild relatives & 2004, Elsevier Ltd. All Rights Reserved. with whom transgenic plantations might repro- duce. Consequently, a major ongoing effort has been sterility transformation that would prevent Introduction completely the sexual reproduction of transgenic plantations. Together with the genera Pinus (subfamily Pinoi- deae), Cathaya, Pseudotsuga (subfamily Laricoideae), See also: Genetics and Genetic Resources: Propaga- and Cedrus, Keteleeria, Nothotsuga, Pseudolarix, tion Technology for Forest Trees. Tree Breeding, Tsuga (subfamily Abietoideae), the genera Picea Practices: Breeding for Disease and Insect Resistance; (subfamily Piceoideae), Abies (subfamily Abietoi- Genetics and Improvement of Wood Properties. Tree deae), and Larix (subfamily Laricoideae) belong to Breeding, Principles: A Historical Overview of Forest the family Pinaceae. The subfamilies are distinguished Tree Improvement; Breeding Theory and Genetic Testing; by cone and seed characters like the existence of an Conifer Breeding Principles and Processes; Economic umbo or the existence of resin vesicles on the seed. Returns from Tree Breeding; Forest Genetics and Tree Species of the genera Picea (spruce), Abies (fir), Breeding; Current and Future Signposts. and Larix (larch) are exclusively distributed in the northern hemisphere from 221 N in the south to 1 Further Reading 73 N forming the polar borderline of trees. Several species of these genera cover wide areas in boreal Dickmann DI and Stuart KW (1983) The Culture of Eurasia and North America. -

Division: SPERMATOPHYTA (Flowering Plants)
Division: SPERMATOPHYTA (Flowering plants) Most important features of plants in Spermtophyta is having flowers and seeds. - Anthophyta (Flowering plants) - Spermatophyta (Plants with seeds) Male gametophyte has become polen grain, and female gametophyte has become embryo vesicle. The seed and the embryo are covered with a special seed coat and can wait for germination until optimal conditions form. Polen grains reach the ovule via wind (anemogamous plants) or bugs (entemogamous plants), etc. Plants are divided into two subclasses as Gymnospermae and Angiospermae according to their ability to form a closed ovarium, or not. anth(o)= Gr. flower; sperm(ato) = Gr. seed gymn(o)= Gr. naked; angio= Gr. narrow (closed) Subdivision: GYMNOSPERMAE (Plants with naked seeds) An ovarium protecting the ovules is not present. Therefore, stylus and stigma are also absent. The micropyle is open and the polen grains enter directly into the polen room found in the ovule and germinate there. Fertilization occurs via the wind, so Gymnospermae plants are anemogamous plants. Gymnospermae plants are in the form of shrubs and trees; herbaceous species are not found among them. Their flowers lack calyx and corolla; male flowers are reduced to polen vesicles and female flowers are reduced to ovules. Both male and female inflorescences are in the form of cones (strobiles). Pollination is via the wind Class: Cycadinae Fam: Cycadaceae Grows in tropical or subtropical regions. Cycas revoluta (King sago, Sago cycad, sikas) A starch called Sago Starch is obtained from this plant and used as food. Class: Ginkgoiae Fam: Ginkgoaceae This family dates back to geological periods, however today it is presented with a single genus and a single species. -
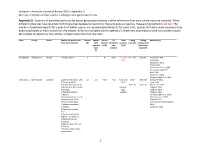
Jacobsen—American Journal of Botany 2021—Appendix S1 Diversity in Conduit and Pit Structure Among Extant Gymnosperm Taxa
Jacobsen—American Journal of Botany 2021—Appendix S1 Diversity in conduit and pit structure among extant gymnosperm taxa Appendix S1. Summary of pit membrane traits for extant gymnosperm genera and the references from which these traits are compiled. When different references have reported conflicting presence/absence reports for the same genus or species, these are highlighted in red text. The number of published reports for a given trait within a genus are included parenthetically for some traits. Species that were newly examined from dried wood blocks or fresh samples for the present review are indicated with an asterisk (*); these new examinations were not counted toward the number of reports nor the numbers of observations for trait characters. Phyla Family Genus Species for which traits Reports Species Vessels Pit Torus Pitting Pitting Reference(s) have been reported (#) with present membrane presence (-seriate) arrangement reported (Y/N) type (Y/N) (alternate/ traits (A/B/C) opposite) (#) Ginkgophyta Ginkgoaceae Ginkgo *Ginkgo biloba L. 9 1 N B (2) Y (1) and uni- & bi- opposite Penhallow 1907, N (6) Liese 1965, Bauch et al. 1972, Timell 1978, Schulte and Gibson 1988, Yin and Xiao-Mei 1992, Dute 1994, Hacke et al. 2004, Schweingruber et al. 2006 Gnetophyta Ephedraceae Ephedra Ephedra americana Humb. 22 12 Y (17) A (2) Y (17) and multi- alternate MacDuffie 1921, & Bonpl. ex Willd., N (2) Liese 1965, Ephedra distachya subsp. (vessels) (vessels) Bauch et al. 1972, helvetica (C.A.Mey.) Asch. (conduit- Carlquist 1990, & Graebn., type Carlquist 1992, Ephedra fasciculata specific) Carlquist 1996, A.Nelson, Schweingruber et al. 2006, Ephedra foeminea Forssk., Motomura et al. -

Sustainable Trees and Shrubs Guide
Sustainable Trees and Shrubs An Introduction to the Sustainable Plant List 1 Planting for Sustainable Landscapes 3 List of Sustainable Trees and Shrubs 8 Index of Common Names 31 Appendix 1: URI Crabapple Tree Disease Evaluations 34 Appendix 2: Tree, Shrub and Vine Selections for Demanding Situations 36 Appendix 3: Cross-reference for Demanding Situations 42 Hardiness Map of New England (USDA Plant Hardiness Zones) 49 An Introduction to the Sustainable Plant List Plant lists are invaluable resources for garden precarious balance of nature in which we play a enthusiasts, designers, nursery trades people and land- pivotal role. Many in our society are looking for ways scape architects. We constantly consult books and to reduce human impacts on the environment. nursery catalogs which list landscape plants, espe- Others lack the time or resources to manage pests or cially those that organize plants by their characteristics maintain shrubs that require frequent pruning or and landscape uses. As times and fashions change, irrigation. These people should find the sustainable new plants emerge, old plants are rediscovered, and plant list an invaluable resource. others lose favor and disappear from these lists. Careful plant selection is the key first step in devel- But one point remains clear: listing plants encourages oping a balanced and self-perpetuating landscape. their widespread distribution and use. The purpose However, plant survival with minimal maintenance is of this publication is to encourage the production not the only issue in sustainability. We are and use of landscape plants that are more sustainable: having more difficulty with invasive those which are not invasive and require reduced exotic plants which inputs of pesticides, water and maintenance. -

Antibacterial Activity of Spruce Bark (Picea Abies L.) Extract Against Escherichia Coli
ABMJ 2018, 1(1): 5-9 DOI: 10.2478/abmj-2018-0001 Acta Biologica Marisiensis ANTIBACTERIAL ACTIVITY OF SPRUCE BARK (PICEA ABIES L.) EXTRACT AGAINST ESCHERICHIA COLI Corneliu TANASE1, Irina BOZ2,3*, Silvia OROIAN1, Sanda COȘARCĂ1, Felicia TOMA1, Anca MARE1, Adrian MAN1 1University of Medicine and Pharmacy of Tîrgu Mureș, Gheorghe Marinescu, 38, 540139, Tîrgu Mureș, Mureș, Romania 2NIRDBS - Institute of Biological Research, Department of Experimental and applied biology, Lascăr Catargi 47,700107, Iași, Romania 3Integrated Centre for Environmental Science Studies in the North-East Development Region – CERNESIM, Alexandru Ioan Cuza University, Carol I 20 A, 700505 Iași, Romania *Correspondence: Irina BOZ [email protected] Received: 17 May 2018; Accepted: 17 June 2018; Published: 30 June 2018 Abstract: The increase of antibiotic resistant bacteria in lasts years resulted in limited options for treatment of bacterial diseases. Escherichia coli is one of the most common Gram-negative bacterial pathogen and a cause of both community and hospital acquired infections. Medicinal plants are alternative rich sources of useful antibacterial agents. The antimicrobial activities of the spruce (Picea abies L.) bark polyphenolic extracts were examined for their capacity to inhibit the growth of Escherichia coli. Spruce bark extract was obtained by conventional aqueous extraction and with ultrasounds. The minimum inhibitory concentration was determined by microdilution method. The antibacterial effect of both extracts was strong against Escherichia coli. The antimicrobial effect of polyphenolic extracts on Escherichia coli was expressed at a concentration of 15 mg/ml. Antimicrobial activity of spruce bark compounds suggest a possible use of spruce bark in pharmaceutical preparations. Keywords: antimicrobial agents, Escherichia coli, polyphenols, spruce bark, Picea abies L.