Susceptibility to Chronic Inflammatory Demyelinating Polyradiculoneuropathy Is Associated to Polymorphic GA Repeat in the SH2D2A Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Validation of a Blood-Based 18-Gene Expression Signature in Colorectal Cancer
Published OnlineFirst March 27, 2013; DOI: 10.1158/1078-0432.CCR-12-3851 Clinical Cancer Imaging, Diagnosis, Prognosis Research Identification and Validation of a Blood-Based 18-Gene Expression Signature in Colorectal Cancer Ye Xu1,4, Qinghua Xu1,6,7, Li Yang1,4, Xun Ye6,7, Fang Liu6,7, Fei Wu6,7, Shujuan Ni1,2,3,5, Cong Tan1,2,3,5, Guoxiang Cai1,4, Xia Meng6,7, Sanjun Cai1,4, and Xiang Du1,2,3,5 Abstract Purpose: The early detection of colorectal cancer (CRC) is crucial for successful treatment and patient survival. However, compliance with current screening methods remains poor. This study aimed to identify an accurate blood-based gene expression signature for CRC detection. Experimental Design: Gene expression in peripheral blood samples from 216 patients with CRC tumors and 187 controls was investigated in the study.Wefirstconductedamicroarrayanalysistoselect candidate genes that were significantly differentially expressed between patients with cancer and con- trols. A quantitative reverse transcription PCR assay was then used to evaluate the expression of selected genes. A gene expression signature was identified using a training set (n ¼ 200) and then validated using an independent test set (n ¼ 160). Results: We identified an 18-gene signature that discriminated the patients with CRC from controls with 92% accuracy, 91% sensitivity, and 92% specificity. The signature performance was further validated in the independent test set with 86% accuracy, 84% sensitivity, and 88% specificity. The area under the receiver operating characteristics curve was 0.94. The signature was shown to be enriched in genes related to immune functions. Conclusions: This study identified an 18-gene signature that accurately discriminated patients with CRC from controls in peripheral blood samples. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Hereditary Duplication of Proximal Chromosome 1Q (Q11q22) in A
1of4 ELECTRONIC LETTER J Med Genet: first published as 10.1136/jmg.39.12.e79 on 1 December 2002. Downloaded from Hereditary duplication of proximal chromosome 1q (q11q22) in a patient with T lymphoblastic lymphoma/leukaemia: a family study using G banding and comparative genomic hybridisation N P H Chan,MHLNg,SHCheng, V Lee, K S Tsang,TTLau,CKLi ............................................................................................................................. J Med Genet 2002;39:e79(http://www.jmedgenet.com/cgi/content/full/39/12/e79) berrations of the long arm of chromosome 1 have been Key points linked to many diseases, both in acquired somatic neo- Aplastic conditions and constitutional genetic disorders. These abnormalities include structural rearrangements and • We report a case of inherited proximal duplication of various deletions or amplifications affecting whole chromo- chromosome 1q in an 11 year old boy presenting with some arms or specific regions. Gain of chromosome 1q has T lymphoblastic lymphoma/leukaemia. The duplication been reported in many solid tumours, including invasive car- of chromosome 1 (q11q22) was identified using G cinomas of the breast, cervical cancers, renal cell carcinoma, banding and confirmed with comparative genomic sarcomas, and hepatocellular carcinomas.1–6 Complete or hybridisation. partial trisomy of 1q has been reported in cases of acute • Cytogenetic studies performed in his family members myeloid leukaemia, myelodysplastic syndrome, myeloprolif- showed the same chromosomal aberration in his mother erative diseases, Fanconi anaemia, and B lineage lymphomas, and sister, but not in his older brother who had a normal where involvement of 1q21 is frequently observed.7–14 karyotype. No dysmorphic syndromes, congenital mal- formations, or psychomotor retardation were observed CASE REPORT in either the index patient or his mother or sister. -

Placental Transcriptome Adaptations to Maternal Nutrient Restriction in Sheep
International Journal of Molecular Sciences Article Placental Transcriptome Adaptations to Maternal Nutrient Restriction in Sheep Chelsie B. Steinhauser 1,†, Colleen A. Lambo 2,†, Katharine Askelson 1, Gregory W. Burns 3, Susanta K. Behura 4,5, Thomas E. Spencer 4, Fuller W. Bazer 1 and Michael Carey Satterfield 1,* 1 Department of Animal Science, Texas A & M University, College Station, TX 77843, USA; [email protected] (C.B.S.); [email protected] (K.A.); [email protected] (F.W.B.) 2 Department of Veterinary Physiology and Pharmacology, Texas A & M University, College Station, TX 77843, USA; [email protected] 3 Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI 49503, USA; [email protected] 4 Division of Animal Sciences, University of Missouri, Columbia, MO 65211, USA; [email protected] (S.K.B.); [email protected] (T.E.S.) 5 Institute for Data Science and Informatics, University of Missouri, Columbia, MO 65211, USA * Correspondence: csatterfi[email protected]; Tel.: +1-979-845-6448 † These authors contributed equally to this work. Abstract: Placental development is modified in response to maternal nutrient restriction (NR), resulting in a spectrum of fetal growth rates. Pregnant sheep carrying singleton fetuses and fed either 100% (n = 8) or 50% (NR; n = 28) of their National Research Council (NRC) recommended intake from days 35–135 of pregnancy were used to elucidate placentome transcriptome alterations at both Citation: Steinhauser, C.B.; Lambo, day 70 and day 135. NR fetuses were further designated into upper (NR NonSGA; n = 7) and lower C.A.; Askelson, K.; Burns, G.W.; quartiles (NR SGA; n = 7) based on day 135 fetal weight. -

1 Novel Expression Signatures Identified by Transcriptional Analysis
ARD Online First, published on October 7, 2009 as 10.1136/ard.2009.108043 Ann Rheum Dis: first published as 10.1136/ard.2009.108043 on 7 October 2009. Downloaded from Novel expression signatures identified by transcriptional analysis of separated leukocyte subsets in SLE and vasculitis 1Paul A Lyons, 1Eoin F McKinney, 1Tim F Rayner, 1Alexander Hatton, 1Hayley B Woffendin, 1Maria Koukoulaki, 2Thomas C Freeman, 1David RW Jayne, 1Afzal N Chaudhry, and 1Kenneth GC Smith. 1Cambridge Institute for Medical Research and Department of Medicine, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK 2Roslin Institute, University of Edinburgh, Roslin, Midlothian, EH25 9PS, UK Correspondence should be addressed to Dr Paul Lyons or Prof Kenneth Smith, Department of Medicine, Cambridge Institute for Medical Research, Addenbrooke’s Hospital, Hills Road, Cambridge, CB2 0XY, UK. Telephone: +44 1223 762642, Fax: +44 1223 762640, E-mail: [email protected] or [email protected] Key words: Gene expression, autoimmune disease, SLE, vasculitis Word count: 2,906 The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in their licence (http://ard.bmj.com/ifora/licence.pdf). http://ard.bmj.com/ on September 29, 2021 by guest. Protected copyright. 1 Copyright Article author (or their employer) 2009. -

By IL-4 in Memory CD8 T Cells Negative Regulation of NKG2D
Negative Regulation of NKG2D Expression by IL-4 in Memory CD8 T Cells Erwan Ventre, Lilia Brinza, Stephane Schicklin, Julien Mafille, Charles-Antoine Coupet, Antoine Marçais, Sophia This information is current as Djebali, Virginie Jubin, Thierry Walzer and Jacqueline of October 2, 2021. Marvel J Immunol published online 31 August 2012 http://www.jimmunol.org/content/early/2012/08/31/jimmun ol.1102954 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2012/09/04/jimmunol.110295 Material 4.DC1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 2, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published August 31, 2012, doi:10.4049/jimmunol.1102954 The Journal of Immunology Negative Regulation of NKG2D Expression by IL-4 in Memory CD8 T Cells Erwan Ventre, Lilia Brinza,1 Stephane Schicklin,1 Julien Mafille, Charles-Antoine Coupet, Antoine Marc¸ais, Sophia Djebali, Virginie Jubin, Thierry Walzer, and Jacqueline Marvel IL-4 is one of the main cytokines produced during Th2-inducing pathologies. -

Agricultural University of Athens
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΩΝ ΖΩΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΖΩΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΝΙΚΗΣ ΚΑΙ ΕΙΔΙΚΗΣ ΖΩΟΤΕΧΝΙΑΣ ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ ΑΘΗΝΑ 2020 ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Genome-wide association analysis and gene network analysis for (re)production traits in commercial broilers ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ Τριμελής Επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Επταμελής εξεταστική επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Πηνελόπη Μπεμπέλη (Καθ. ΓΠΑ) Δημήτριος Βλαχάκης (Επ. Καθ. ΓΠΑ) Ευάγγελος Ζωίδης (Επ.Καθ. ΓΠΑ) Γεώργιος Θεοδώρου (Επ.Καθ. ΓΠΑ) 2 Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Περίληψη Σκοπός της παρούσας διδακτορικής διατριβής ήταν ο εντοπισμός γενετικών δεικτών και υποψηφίων γονιδίων που εμπλέκονται στο γενετικό έλεγχο δύο τυπικών πολυγονιδιακών ιδιοτήτων σε κρεοπαραγωγικά ορνίθια. Μία ιδιότητα σχετίζεται με την ανάπτυξη (σωματικό βάρος στις 35 ημέρες, ΣΒ) και η άλλη με την αναπαραγωγική -

PP1-Associated Signaling and − B/AP-1 Κ Inhibition of NF- Tolerance
Downloaded from http://www.jimmunol.org/ by guest on October 3, 2021 is online at: average * and − B/AP-1 κ The Journal of Immunology published online 26 February 2014 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2014/02/26/jimmun ol.1301610 Identification of Two Forms of TNF Tolerance in Human Monocytes: Differential Inhibition of NF- PP1-Associated Signaling Johannes Günther, Nico Vogt, Katharina Hampel, Rolf Bikker, Sharon Page, Benjamin Müller, Judith Kandemir, Michael Kracht, Oliver Dittrich-Breiholz, René Huber and Korbinian Brand J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2014/02/26/jimmunol.130161 0.DCSupplemental Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 3, 2021. Published February 26, 2014, doi:10.4049/jimmunol.1301610 The Journal of Immunology Identification of Two Forms of TNF Tolerance in Human Monocytes: Differential Inhibition of NF-kB/AP-1– and PP1-Associated Signaling Johannes Gunther,*€ ,1 Nico Vogt,*,1 Katharina Hampel,*,1 Rolf Bikker,* Sharon Page,* Benjamin Muller,*€ Judith Kandemir,* Michael Kracht,† Oliver Dittrich-Breiholz,‡ Rene´ Huber,* and Korbinian Brand* The molecular basis of TNF tolerance is poorly understood. -
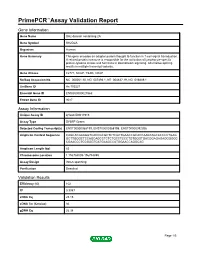
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name SH2 domain containing 2A Gene Symbol SH2D2A Organism Human Gene Summary This gene encodes an adaptor protein thought to function in T-cell signal transduction. A related protein in mouse is responsible for the activation of lymphocyte-specific protein-tyrosine kinase and functions in downstream signaling. Alternative splicing results in multiple transcript variants. Gene Aliases F2771, SCAP, TSAD, VRAP RefSeq Accession No. NC_000001.10, NG_007493.1, NT_004487.19, NG_016849.1 UniGene ID Hs.103527 Ensembl Gene ID ENSG00000027869 Entrez Gene ID 9047 Assay Information Unique Assay ID qHsaCID0011915 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000368199, ENST00000368198, ENST00000392306 Amplicon Context Sequence CAGCACGAAGGTCACCGCGCTCTCGCTGAACCGCACCAAGTAGCACCCCTGAG GCTTGGGCTCCAGCAGCCTCTCTGCCTCCCTGTGGGTGACGGAGAGAGGGGGC CGAACCCTCCGGGTGATGAAGCCATGGAACCAGGCAG Amplicon Length (bp) 83 Chromosome Location 1:156784005-156784859 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 102 R2 0.9987 cDNA Cq 25.15 cDNA Tm (Celsius) 86 gDNA Cq 26.38 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SH2D2A, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 3/5 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein -
T Cell–Specific Adaptor Protein Regulates Mitochondrial Function
Published September 20, 2019, doi:10.4049/jimmunol.1801604 The Journal of Immunology T Cell–Specific Adaptor Protein Regulates Mitochondrial Function and CD4+ T Regulatory Cell Activity In Vivo following Transplantation Johannes Wedel,*,†,‡ Maria P. Stack,*,†,‡ Tatsuichiro Seto,*,†,‡ Matthew M. Sheehan,*,† Evelyn A. Flynn,*,†,‡ Isaac E. Stillman,x,{ Sek Won Kong,‡,‖ Kaifeng Liu,*,†,‡,# and David M. Briscoe*,†,‡ The T cell–specific adaptor protein (TSAd), encoded by the SH2D2A gene, is an intracellular molecule that binds Lck to elicit signals that result in cytokine production in CD4+ T effector cells (Teff). Nevertheless, using Sh2d2a knockout (KO; also called TSAd2/2) mice, we find that alloimmune CD4+ Teff responses are fully competent in vivo. Furthermore, and contrary to expectations, we find that allograft rejection is accelerated in KO recipients of MHC class II–mismatched B6.C-H-2bm12 heart transplants versus wild-type (WT) recipients. Also, KO recipients of fully MHC-mismatched cardiac allografts are resistant to the graft-prolonging effects of costimulatory blockade. Using adoptive transfer models, we find that KO T regulatory cells (Tregs) are less efficient in suppressing Teff function and they produce IFN-g following mitogenic activation. In addition, pyrosequencing demonstrated higher levels of methylation of CpG regions within the Treg-specific demethylated region of KO versus WT Tregs, suggesting that TSAd, in part, promotes Treg stability. By Western blot, Lck is absent in the mitochondria of KO Tregs, and reactive oxygen species production by mitochondria is reduced in KO versus WT Tregs. Full transcriptomic analysis demonstrated that the key mechanism of TSAd function in Tregs relates to its effects on cellular activation rather than intrinsic effects on mitochondria/metabolism. -
Chromosome 12, Frequently Deleted in Human Pancreatic Cancer, May Encode a Tumor-Suppressor Gene That Suppresses Angiogenesis
Laboratory Investigation (2004) 84, 1339–1351 & 2004 USCAP, Inc All rights reserved 0023-6837/04 $30.00 www.laboratoryinvestigation.org Chromosome 12, frequently deleted in human pancreatic cancer, may encode a tumor-suppressor gene that suppresses angiogenesis Sumitaka Yamanaka1,2,*, Makoto Sunamura3,*, Toru Furukawa1, Libo Sun3, Liviu P Lefter1,3, Tadayoshi Abe1,3, Toshimasa Yatsuoka1,3, Hiroko Fujimura3, Emiko Shibuya3, Noriko Kotobuki4, Mitsuo Oshimura4, Akira Sakurada2, Masami Sato2, Takashi Kondo2, Seiki Matsuno3 and Akira Horii1 1Department of Molecular Pathology, Tohoku University School of Medicine, Sendai, Japan; 2Department of Thoracic Surgery, Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan; 3Department of Gastroenterological Surgery, Tohoku University School of Medicine, Sendai, Japan and 4Department of Cell Technology, Tottori University School of Medicine, Yonago, Japan Several lines of evidence have suggested that the long arm of chromosome 12 may carry a tumor-suppressor gene(s) that plays a role in pancreatic ductal carcinogenesis. We have previously found a significant association between loss of heterozygosity of the 12q arm and a poor prognosis in pancreatic cancer patients. In this study, we introduced a normal copy of chromosome 12 into some pancreatic ductal carcinoma cells. Both anchorage-dependent and -independent proliferations as well as invasiveness were similar throughout the hybrid clones when compared with their corresponding parental cells. In sharp contrast, significant suppression of tumorigenesis was observed after inoculation of the hybrid clones into nude mice. Measurements made up to 1 month later showed that there was a significant delay in the growth of tumors into which the introduced normal copy of chromosome 12 had been restored.