Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Art Gallery of Ballarat Annual Report 10-11 Annual Report
Art Gallery of Ballarat Annual Report 10-11 Annual Report 2010-11 ISSN 0726-5530 Chair’s Report .................................................................................................4 Art Gallery of Ballarat ACN: 145 246 224 Director’s Report .........................................................................................6 ABN: 28 145 246 224 Association Report .....................................................................................8 40 Lydiard Street North Ballarat Victoria 3350 Women’s Association Report ............................................................10 T 03 5320 5858 F 03 5320 5791 Gallery Guides Report ...........................................................................11 [email protected] Acquisitions ...................................................................................................13 www.artgalleryofballarat.com.au Outward Loan ..............................................................................................27 Exhibitions ......................................................................................................31 Public Programs ........................................................................................35 Education Visits and Programs ..........................................................37 Adopt an Artwork ......................................................................................40 Donations, Gifts and Bequests .........................................................41 Gallery Staff and Volunteers -
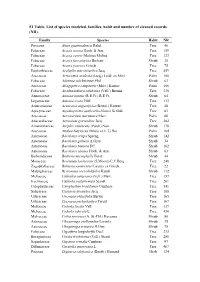
S1 Table. List of Species Modeled, Families, Habit and Number of Cleaned Records (NR)
S1 Table. List of species modeled, families, habit and number of cleaned records (NR). Family Species Habit NR Pinaceae Abies guatemalensis Rehd. Tree 46 Fabaceae Acacia aroma Hook. & Arn. Tree 159 Fabaceae Acacia caven (Molina) Molina Tree 123 Fabaceae Acacia furcatispina Burkart Shrub 35 Fabaceae Acacia praecox Griseb. Tree 75 Euphorbiaceae Acalypha macrostachya Jacq. Tree 459 Arecaceae Acrocomia aculeata (Jacq.) Lodd. ex Mart. Palm 166 Fabaceae Adesmia volckmannii Phil. Shrub 63 Arecaceae Allagoptera campestris (Mart.) Kuntze Palm 106 Fabaceae Anadenanthera colubrina (Vell.) Brenan Tree 330 Annonaceae Annona nutans (R.E.Fr.) R.E.Fr. Shrub 64 Loganiaceae Antonia ovata Pohl Tree 113 Araucariaceae Araucaria angustifolia (Bertol.) Kuntze Tree 48 Apocynaceae Aspidosperma quebracho-blanco Schltdl. Tree 83 Arecaceae Astrocaryum murumuru Mart. Palm 66 Anacardiaceae Astronium graveolens Jacq. Tree 282 Amaranthaceae Atriplex canescens (Pursh) Nutt. Shrub 178 Arecaceae Attalea butyracea (Mutis ex L.f.) We Palm 104 Asteraceae Baccharis crispa Spreng. Shrub 142 Asteraceae Baccharis gilliesii A.Gray Shrub 34 Asteraceae Baccharis trimera DC. Shrub 162 Asteraceae Baccharis ulicina Hook. & Arn. Shrub 63 Berberidaceae Berberis microphylla Forst. Shrub 44 Moraceae Brosimum lactescens (S.Moore) C.C.Berg Tree 248 Zygophyllaceae Bulnesia sarmientoi Lorentz ex Griseb. Tree 22 Malpighiaceae Byrsonima coccolobifolia Kunth Shrub 112 Meliaceae Cabralea canjerana (Vell.) Mart. Tree 157 Icacinaceae Calatola costaricensis Standl. Tree 201 Calophyllaceae Calophyllum brasiliense Cambess. Tree 541 Salicaceae Casearia decandra Jacq. Tree 188 Urticaceae Cecropia obtusifolia Bertol. Tree 165 Urticaceae Cecropia pachystachya Trécul Tree 167 Meliaceae Cedrela fissilis Vell. Tree 137 Meliaceae Cedrela odorata L. Tree 436 Malvaceae Ceiba speciosa (A. St.-Hil.) Ravenna Shrub 80 Asteraceae Chuquiraga avellanedae Lorentz Shrub 35 Asteraceae Chuquiraga erinacea D.Don Shrub 75 Fabaceae Copaifera langsdorffii Desf. -

Supplementary Materialsupplementary Material
10.1071/BT13149_AC © CSIRO 2013 Australian Journal of Botany 2013, 61(6), 436–445 SUPPLEMENTARY MATERIAL Comparative dating of Acacia: combining fossils and multiple phylogenies to infer ages of clades with poor fossil records Joseph T. MillerA,E, Daniel J. MurphyB, Simon Y. W. HoC, David J. CantrillB and David SeiglerD ACentre for Australian National Biodiversity Research, CSIRO Plant Industry, GPO Box 1600 Canberra, ACT 2601, Australia. BRoyal Botanic Gardens Melbourne, Birdwood Avenue, South Yarra, Vic. 3141, Australia. CSchool of Biological Sciences, Edgeworth David Building, University of Sydney, Sydney, NSW 2006, Australia. DDepartment of Plant Biology, University of Illinois, Urbana, IL 61801, USA. ECorresponding author. Email: [email protected] Table S1 Materials used in the study Taxon Dataset Genbank Acacia abbreviata Maslin 2 3 JF420287 JF420065 JF420395 KC421289 KC796176 JF420499 Acacia adoxa Pedley 2 3 JF420044 AF523076 AF195716 AF195684; AF195703 Acacia ampliceps Maslin 1 KC421930 EU439994 EU811845 Acacia anceps DC. 2 3 JF420244 JF420350 JF419919 JF420130 JF420456 Acacia aneura F.Muell. ex Benth 2 3 JF420259 JF420036 JF420366 JF419935 JF420146 KF048140 Acacia aneura F.Muell. ex Benth. 1 2 3 JF420293 JF420402 KC421323 JQ248740 JF420505 Acacia baeuerlenii Maiden & R.T.Baker 2 3 JF420229 JQ248866 JF420336 JF419909 JF420115 JF420448 Acacia beckleri Tindale 2 3 JF420260 JF420037 JF420367 JF419936 JF420147 JF420473 Acacia cochlearis (Labill.) H.L.Wendl. 2 3 KC283897 KC200719 JQ943314 AF523156 KC284140 KC957934 Acacia cognata Domin 2 3 JF420246 JF420022 JF420352 JF419921 JF420132 JF420458 Acacia cultriformis A.Cunn. ex G.Don 2 3 JF420278 JF420056 JF420387 KC421263 KC796172 JF420494 Acacia cupularis Domin 2 3 JF420247 JF420023 JF420353 JF419922 JF420133 JF420459 Acacia dealbata Link 2 3 JF420269 JF420378 KC421251 KC955787 JF420485 Acacia dealbata Link 2 3 KC283375 KC200761 JQ942686 KC421315 KC284195 Acacia deanei (R.T.Baker) M.B.Welch, Coombs 2 3 JF420294 JF420403 KC421329 KC955795 & McGlynn JF420506 Acacia dempsteri F.Muell. -

Martu Aboriginal People's Uses and Knowledge of Their
To hunt and to hold: Martu Aboriginal people’s uses and knowledge of their country, with implications for co-management in Karlamilyi (Rudall River) National Park and the Great Sandy Desert, Western Australia Fiona J. Walsh, B.Sc. (Zoology), M.Sc. Prelim. (Botany) This thesis is presented for the degree of Doctor of Philosophy of The University of Western Australia School of Social and Cultural Studies (Anthropology) and School of Plant Biology (Ecology) 2008 i Photo i (title page) Rita Milangka displays Lil-lilpa (Fimbristylis eremophila), the UWA Department of Botany field research vehicle is in background. This sedge has numerous small seeds that were ground into an edible paste. Whilst Martu did not consume sedge and grass seeds in contemporary times, their use was demonstrated to younger people and visitors. ii DEDICATION This dissertation is dedicated to my parents, Dianne and John Walsh. My mother cultivated my joy in plants and wildlife. She introduced me to my first bush foods (including kurarra, kogla, ‘honeysuckle’, bardi grubs) on Murchison lands inhabited by Yamatji people then my European pastoralist forbearers.1 My father shares bush skills, a love of learning and long stories. He provided his Toyota vehicle and field support to me on Martu country in 1988. The dedication is also to Martu yakurti (mothers) and mama (fathers) who returned to custodial lands to make safe homes for children and their grandparents and to hold their country for those past and future generations. Photo ii John, Dianne and Melissa Walsh (right to left) net for Gilgie (Freshwater crayfish) on Murrum in the Murchison. -

(Alhagi Maurorum) As New Sources of Bioactive Compounds Using GC-MS Technique
POJ 12(01):70-77 (2019) ISSN:1836-3644 doi: 10.21475/poj.12.01.19.pt1971 Assessment of camel thorn (Alhagi maurorum) as new sources of bioactive compounds using GC-MS technique Reham M. Mostafa, Heba S. Essawy* Botany Department, Facukty of Science, Benha University, Egypt Corresponding author: [email protected] Abstract Alhagi maurorum (A. maurorum) is one of the medicinally important plants belonging to the family leguminasae, commonly known as camel thorn. This research was amid to identify the chemical compounds in the aerial part of A. maurorum using GC-mass analysis. Three solvents with different polarities were used for the extraction of chemical constituents (water, methanol and petroleum ether). The results of GC-MS analysis led to identification of various compounds. In total, thirty-nine compounds from petroleum ether extract, thirty-two compounds in methanolic extract and seventeen compounds in aqueous extract were identified. Majority of the identified compounds have been reported to possess many biological activities. Among them, we reported 10 new anticancer compounds (Vitamin E; Hexadecanoic acid; Stigmast-5-en-3-ol; Phytol,2-hexadecen-1-ol,3,7,11,15-tetramethyl; Squalene; Hexadecanoic acid; 2-hydroxy-1-(hydroxymethyl) ethyl ester; Oxime,methoxy-phenyl,methyl N-hydroxyben- zenecarboximidoate; Ergost-5-en-3-ol; 9,12- Octadecad-ienoic acid and Farnesol) from A. maurorum using three solvent, while the best effective solvent was petroluem ether. Therefore, we report that A. maurorum has great potential to be developed into anticancer drugs. Keywords: Secondary metabolites; Alhagi maurarum; GC-mass spectroscopy; anticancer compounds. Introduction Natural products have useful and interesting biological tissue-repairing properties (Al-Snafi, 2015). -

ISTA List of Stabilized Plant Names 7Th Edition
ISTA List of Stabilized Plant Names th 7 Edition ISTA Nomenclature Committee Chair: Dr. M. Schori Published by All rights reserved. No part of this publication may be The Internation Seed Testing Association (ISTA) reproduced, stored in any retrieval system or transmitted Zürichstr. 50, CH-8303 Bassersdorf, Switzerland in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior ©2020 International Seed Testing Association (ISTA) permission in writing from ISTA. ISBN 978-3-906549-77-4 ISTA List of Stabilized Plant Names 1st Edition 1966 ISTA Nomenclature Committee Chair: Prof P. A. Linehan 2nd Edition 1983 ISTA Nomenclature Committee Chair: Dr. H. Pirson 3rd Edition 1988 ISTA Nomenclature Committee Chair: Dr. W. A. Brandenburg 4th Edition 2001 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 5th Edition 2007 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 6th Edition 2013 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 7th Edition 2019 ISTA Nomenclature Committee Chair: Dr. M. Schori 2 7th Edition ISTA List of Stabilized Plant Names Content Preface .......................................................................................................................................................... 4 Acknowledgements ....................................................................................................................................... 6 Symbols and Abbreviations .......................................................................................................................... -

Tree and Tree-Like Species of Mexico: Asteraceae, Leguminosae, and Rubiaceae
Revista Mexicana de Biodiversidad 84: 439-470, 2013 Revista Mexicana de Biodiversidad 84: 439-470, 2013 DOI: 10.7550/rmb.32013 DOI: 10.7550/rmb.32013439 Tree and tree-like species of Mexico: Asteraceae, Leguminosae, and Rubiaceae Especies arbóreas y arborescentes de México: Asteraceae, Leguminosae y Rubiaceae Martin Ricker , Héctor M. Hernández, Mario Sousa and Helga Ochoterena Herbario Nacional de México, Departamento de Botánica, Instituto de Biología, Universidad Nacional Autónoma de México. Apartado postal 70- 233, 04510 México D. F., Mexico. [email protected] Abstract. Trees or tree-like plants are defined here broadly as perennial, self-supporting plants with a total height of at least 5 m (without ascending leaves or inflorescences), and with one or several erect stems with a diameter of at least 10 cm. We continue our compilation of an updated list of all native Mexican tree species with the dicotyledonous families Asteraceae (36 species, 39% endemic), Leguminosae with its 3 subfamilies (449 species, 41% endemic), and Rubiaceae (134 species, 24% endemic). The tallest tree species reach 20 m in the Asteraceae, 70 m in the Leguminosae, and also 70 m in the Rubiaceae. The species-richest genus is Lonchocarpus with 67 tree species in Mexico. Three legume genera are endemic to Mexico (Conzattia, Hesperothamnus, and Heteroflorum). The appendix lists all species, including their original publication, references of taxonomic revisions, existence of subspecies or varieties, maximum height in Mexico, and endemism status. Key words: biodiversity, flora, tree definition. Resumen. Las plantas arbóreas o arborescentes se definen aquí en un sentido amplio como plantas perennes que se pueden sostener por sí solas, con una altura total de al menos 5 m (sin considerar hojas o inflorescencias ascendentes) y con uno o varios tallos erectos de un diámetro de al menos 10 cm. -

Flora Survey on Hiltaba Station and Gawler Ranges National Park
Flora Survey on Hiltaba Station and Gawler Ranges National Park Hiltaba Pastoral Lease and Gawler Ranges National Park, South Australia Survey conducted: 12 to 22 Nov 2012 Report submitted: 22 May 2013 P.J. Lang, J. Kellermann, G.H. Bell & H.B. Cross with contributions from C.J. Brodie, H.P. Vonow & M. Waycott SA Department of Environment, Water and Natural Resources Vascular plants, macrofungi, lichens, and bryophytes Bush Blitz – Flora Survey on Hiltaba Station and Gawler Ranges NP, November 2012 Report submitted to Bush Blitz, Australian Biological Resources Study: 22 May 2013. Published online on http://data.environment.sa.gov.au/: 25 Nov. 2016. ISBN 978-1-922027-49-8 (pdf) © Department of Environment, Water and Natural Resouces, South Australia, 2013. With the exception of the Piping Shrike emblem, images, and other material or devices protected by a trademark and subject to review by the Government of South Australia at all times, this report is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. All other rights are reserved. This report should be cited as: Lang, P.J.1, Kellermann, J.1, 2, Bell, G.H.1 & Cross, H.B.1, 2, 3 (2013). Flora survey on Hiltaba Station and Gawler Ranges National Park: vascular plants, macrofungi, lichens, and bryophytes. Report for Bush Blitz, Australian Biological Resources Study, Canberra. (Department of Environment, Water and Natural Resources, South Australia: Adelaide). Authors’ addresses: 1State Herbarium of South Australia, Department of Environment, Water and Natural Resources (DEWNR), GPO Box 1047, Adelaide, SA 5001, Australia. -

Pilbara 1 (PIL1 – Chichester Subregion)
Pilbara 1 Pilbara 1 (PIL1 – Chichester subregion) PETER KENDRICK AND NORM MCKENZIE AUGUST 2001 Subregional description and biodiversity arnhemensis and other Critical Weight Range mammals, arid zone populations of Ghost Bat (Macroderma gigas), values Northwestern Long-eared Bat (Nyctophilus bifax daedalus) and Little Northwestern Free-tailed Bat Description and area (Mormopterus loriae cobourgensis) are also significant in the subregion. The Chichester subregion (PIL 1) comprises the northern section of the Pilbara Craton. Undulating Rare Flora: Archaean granite and basalt plains include significant Species of subregional significance include Livistona areas of basaltic ranges. Plains support a shrub steppe alfredii populations in the Chichester escarpment characterised by Acacia inaequilatera over Triodia (Sherlock River drainage). wiseana (formerly Triodia pungens) hummock grasslands, while Eucalyptus leucophloia tree steppes occur on ranges. Centres of endemism: The climate is Semi-desert-tropical and receives 300mm Bioregional endemics include Ningaui timealeyi, an of rainfall annually. Drainage occurs to the north via undescribed Planigale, Dasykaluta rosamondae, numerous rivers (e.g. De Grey, Oakover, Nullagine, Pseudomys chapmani, Pseudantechinus roryi, Diplodactylus Shaw, Yule, Sherlock). Subregional area is 9,044,560ha. savagei, Diplodactylus wombeyi, Delma elegans, Delma pax, Ctenotus rubicundus, Ctenotus affin. robustus, Egernia pilbarensis, Lerista zietzi, Lerista flammicauda, Dominant land use Lerista neander, two or three undescribed taxa within Lerista muelleri, Notoscincus butleri, Varanus pilbarensis, Grazing – native pastures (see Appendix B, key b), Acanthophis wellsi, Demansia rufescens, Ramphotyphlops Aboriginal lands and Reserves, UCL & Crown Reserves, pilbarensis, and Ramphotyphlops ganei. Conservation, and Mining leases. Refugia: Continental Stress Class There are no known true Refugia in PIL1, however it is possible that calcrete aquifers in the upper Oakover Continental Stress Class for PIL1 is 4. -

Boletín Del Instituto De Botánica
ISSN 0187-7054 muG BOLETÍN DEL INSTITUTO DE BOTÁNICA Vol. 8 Núm. 1-2 8 de noviembre de 2000 Fecha efectiva de publicación 3 de abril de 2001 CUCBA UNIVERSIDAD DE GUADALAJARA RECTORÍA GENERAL DEPARTAMENTO DE BOTÁNICA Y ZOOLOGÍA Dr. Víctor Manuel González Romero Rector Dr. J. Antonio V ázquez García Jefe del Departamento Dr. Misael Gradilla Damy Vicerrector Ejecutivo INSTITUTO DE BOTÁNICA Lic. J. Trinidad Padilla López COMITÉ EDITORIAL Secretario General CENTRO UNIVERSITARIO Roberto González Tamayo DE CIENCIAS BIOLÓGICAS Coordinador de edición Y AGROPECUARIAS Adriana Patricia Miranda Núñez M. en C. Salvador Mena Munguía Responsable de edición Rector . Servando Carvajal H . M. en C. Santiago Sánchez Preciado Secretario Académico Laura Guzmán Dávalos M.V.Z. José Rizo Ayala Mollie Harker de Rodríguez Secretario Administrativo Jorge A. Pérez de la Rosa DIVISIÓN DE CIENCIAS' BIOLÓ- J. Jacqueline Reynoso Dueñas GICAS Y AMBIENTALES J. Antonio Vázquez García Dr. Arturo Orozco Barocio Director Luz Ma. Villarreal de Puga M. en C. Martha Georgina Orozco Medina Secretario Fecha efectiva de publicación 3 de abril de 2001 ~~1!}J ! 8 u<!;; CONTENIDO lft,\ lS~.:o r.... ~;tib)~~- LAS ESPECIES JALISCIENSES DEL GÉNERO FICUS L. (MORACEAE) .............. .................................................... Roberto Quintana-Cardoza y Servando Carvajal! MORFOLOGÍA DEL POLEN DE AMPHIPTERYGIUM SCHIEDE ex STANDLEY (JULIANIACEAE) •••••• Noemí Jiménez-Reyes y Xochitl Marisol Cuevas-Figueroa 65 FLORÍSTICA DEL CERRO DEL COLLI, MUNICIPIO DE ZAPOPAN, JALISCO, MÉXICO ............. Miguel A. Macias-Rodríguez y Raymundo Ramírez-Delgadillo 75 ESTUDIO PALINOLÓGICO DE ESPECIES DEL GÉNERO POPULUS L. (SALICACEAE) EN MÉXICO ................................................................................. .... .. .. .. .... .. ...... .. .. ... .. .. .. Rosa Elena Martínez-González y Noemí Jiménez-Reyes 1O 1 COMUNIDADES DE MACROALGAS EN AMBIENTES INTERMAREALES DEL SURESTE DE BAHÍA TENACATITA, JALISCO, MÉXICO ................................ -

Synoptic Overview of Exotic Acacia, Senegalia and Vachellia (Caesalpinioideae, Mimosoid Clade, Fabaceae) in Egypt
plants Article Synoptic Overview of Exotic Acacia, Senegalia and Vachellia (Caesalpinioideae, Mimosoid Clade, Fabaceae) in Egypt Rania A. Hassan * and Rim S. Hamdy Botany and Microbiology Department, Faculty of Science, Cairo University, Giza 12613, Egypt; [email protected] * Correspondence: [email protected] Abstract: For the first time, an updated checklist of Acacia, Senegalia and Vachellia species in Egypt is provided, focusing on the exotic species. Taking into consideration the retypification of genus Acacia ratified at the Melbourne International Botanical Congress (IBC, 2011), a process of reclassification has taken place worldwide in recent years. The review of Acacia and its segregates in Egypt became necessary in light of the available information cited in classical works during the last century. In Egypt, various taxa formerly placed in Acacia s.l., have been transferred to Acacia s.s., Acaciella, Senegalia, Parasenegalia and Vachellia. The present study is a contribution towards clarifying the nomenclatural status of all recorded species of Acacia and its segregate genera. This study recorded 144 taxa (125 species and 19 infraspecific taxa). Only 14 taxa (four species and 10 infraspecific taxa) are indigenous to Egypt (included now under Senegalia and Vachellia). The other 130 taxa had been introduced to Egypt during the last century. Out of the 130 taxa, 79 taxa have been recorded in literature. The focus of this study is the remaining 51 exotic taxa that have been traced as living species in Egyptian gardens or as herbarium specimens in Egyptian herbaria. The studied exotic taxa are accommodated under Acacia s.s. (24 taxa), Senegalia (14 taxa) and Vachellia (13 taxa). -

Species Summary
Acacia jennerae LC Taxonomic Authority: Maiden Global Assessment Regional Assessment Region: Global Endemic to region Synonyms Common Names Racosperma jennera (Maiden) Pedley COONAVITTRA WATTLE English Upper Level Taxonomy Kingdom: PLANTAE Phylum: TRACHEOPHYTA Class: MAGNOLIOPSIDA Order: FABALES Family: LEGUMINOSAE Lower Level Taxonomy Rank: Infra- rank name: Plant Hybrid Subpopulation: Authority: In the past the species has commonly been confused with Acacia microbotrya; their ranges overlap in the Burakin–Trayning area in Western Australia. A. jennerae is normally distinguished by its straighter phyllodes with more numerous glands and its funicle (Orchard and Wilson 2001). General Information Distribution Acacia jennerae is endemic to Australia, distributed from near Kununoppin in Western Australia, through South Australia and the Northern Territory to Wilcannia in New South Wales, and the Simpson Desert in far south-west Queensland (Orchard and Wilson 2001). Range Size Elevation Biogeographic Realm Area of Occupancy: Upper limit: 900 Afrotropical Extent of Occurrence: Lower limit: 60 Antarctic Map Status: Depth Australasian Upper limit: Neotropical Lower limit: Oceanian Depth Zones Palearctic Shallow photic Bathyl Hadal Indomalayan Photic Abyssal Nearctic Population Total population size is not known but it was recently collected in 2002. Total Population Size Minimum Population Size: Maximum Population Size: Habitat and Ecology A tall shrub or tree that has the habit of a small mallee eucalypt that is sporadic in arid and semi-arid areas. It grows in mallee and savannah woodland. System Movement pattern Crop Wild Relative Terrestrial Freshwater Nomadic Congregatory/Dispersive Is the species a wild relative of a crop? Marine Migratory Altitudinally migrant Growth From Definition Shrub - large Perennial shrub (>1m), also termed a Phanerophyte (>1m) Tree - large Large tree, also termed a Phanerophyte (>1m) Threats There are no major threats known to this widespread species.