Coordinated Regulation of Gene Expression by E.Coli Chromatin Proteins FIS and H-NS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Recruitment of Genes and Enzymes Conferring Resistance to the Nonnatural Toxin Bromoacetate
Recruitment of genes and enzymes conferring resistance to the nonnatural toxin bromoacetate Kevin K. Desai and Brian G. Miller1 Department of Chemistry and Biochemistry, Florida State University, Tallahassee, FL 32306-4390 Edited* by Richard Wolfenden, University of North Carolina, Chapel Hill, NC, and approved August 24, 2010 (received for review May 28, 2010) Microbial niches contain toxic chemicals capable of forcing organ- tance of a naïve bacterial population can play a role in combating isms into periods of intense natural selection to afford survival. the toxicity of a nonnatural small-molecule. Revealing the reser- Elucidating the mechanisms by which microbes evade environmen- voir of intrinsic resistance genes that are subject to evolutionary tal threats has direct relevance for understanding and combating recruitment promises to aid our understanding of the processes the rise of antibiotic resistance. In this study we used a toxic small- leading to the emergence of antibiotic resistant pathogens. molecule, bromoacetate, to model the selective pressures imposed We sought to identify the full spectrum of bromoacetate resis- by antibiotics and anthropogenic toxins. We report the results tance mechanisms available to the model bacterium, Escherichia of genetic selection experiments that identify nine genes from coli. The reactivity of bromoacetate is likely to mimic that of elec- Escherichia coli whose overexpression affords survival in the trophilic natural products as well as anthropogenic environmen- presence of a normally lethal concentration of bromoacetate. Eight tal contaminants that microbes may encounter. The clinically of these genes encode putative transporters or transmembrane significant natural antibiotic fosfomycin, and the fungal natural proteins, while one encodes the essential peptidoglycan biosyn- product terreic acid, are electrophilic molecules that both target N thetic enzyme, UDP- -acetylglucosamine enolpyruvoyl transferase an essential nucleophilic cysteine residue in bacteria (8, 9). -
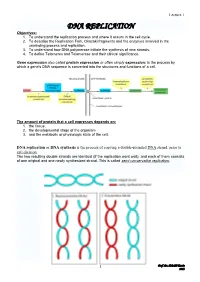
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

De Novo Sequencing and Transcriptome Analysis Reveal Key Genes Regulating Steroid Metabolism in Leaves, Roots, Adventitious Roots and Calli of Periploca Sepium Bunge
ORIGINAL RESEARCH published: 21 April 2017 doi: 10.3389/fpls.2017.00594 De novo Sequencing and Transcriptome Analysis Reveal Key Genes Regulating Steroid Metabolism in Leaves, Roots, Adventitious Roots and Calli of Periploca sepium Bunge Jian Zhang 1, 2, 3, Xinglin Li 1, 3*, Fuping Lu 1, 3, Shanying Wang 1, 3, Yunhe An 4, Xiaoxing Su 4, Xiankuan Li 2, Lin Ma 2 and Guangjian Han 5 1 Key Lab of Industrial Fermentation Microbiology, Tianjin University of Science and Technology, Ministry of Education, Tianjin, China, 2 School of Traditional Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, Tianjin, China, 3 College of Bioengineering, Tianjin University of Science and Technology, Tianjin, China, 4 Beijing Center for Physical and Chemical Analysis, Beijing, China, 5 Shachuan Biotechnology, Tianjin, China Edited by: Periploca sepium Bunge is a traditional medicinal plant, whose root bark is important Peng Zhang, Institute of Plant Physiology and for Chinese herbal medicine. Its major bioactive compounds are C21 steroids and Ecology, SIBS, CAS, China periplocin, a kind of cardiac glycoside, which are derived from the steroid synthesis Reviewed by: pathway. However, research on P. sepium genome or transcriptomes and their related Kun Yu, genes has been lacking for a long time. In this study we estimated this species Hubei University of Chinese Medicine, China nuclear genome size at 170 Mb (using flow cytometry). Then, RNA sequencing of Jun Yang, four different tissue samples of P. sepium (leaves, roots, adventitious roots, and Shanghai Chenshan Plant Science Research Center (CAS), China calli) was done using the sequencing platform Illumina/Solexa Hiseq 2,500. -

The Cyanide Hydratase from Neurospora Crassa Forms a Helix Which Has a Dimeric Repeat
Appl Microbiol Biotechnol (2009) 82:271–278 DOI 10.1007/s00253-008-1735-4 BIOTECHNOLOGICALLY RELEVANT ENZYMES AND PROTEINS The cyanide hydratase from Neurospora crassa forms a helix which has a dimeric repeat Kyle C. Dent & Brandon W. Weber & Michael J. Benedik & B. Trevor Sewell Received: 2 July 2008 /Revised: 24 September 2008 /Accepted: 25 September 2008 / Published online: 23 October 2008 # Springer-Verlag 2008 Abstract The fungal cyanide hydratases form a functionally Introduction specialized subset of the nitrilases which catalyze the hydrolysis of cyanide to formamide with high specificity. The generation of cyanide containing waste results from These hold great promise for the bioremediation of cyanide cyanide utilization in a variety of applications, spanning wastes. The low resolution (3.0 nm) three-dimensional such diverse industries as gold and silver mining, metal reconstruction of negatively stained recombinant cyanide electroplating, polymer synthesis, and the production of hydratase fibers from the saprophytic fungus Neurospora fine chemicals, pharmaceuticals, dyes, and agricultural crassa by iterative helical real space reconstruction reveals products (Baxter and Cummings 2006). Traditionally, that enzyme fibers display left-handed D1 S5.4 symmetry chemical oxidation or stabilizations are used to treat these with a helical rise of 1.36 nm. This arrangement differs from wastes; however, many groups have been studying the previously characterized microbial nitrilases which demon- potential for bioremediation of such cyanide waste strate a structure built along similar principles but with a (O’Reilly and Turner 2003; Jandhyala et al. 2005). reduced helical twist. The cyanide hydratase assembly is The most promising approach utilizes cyanide degrada- stabilized by two dyadic interactions between dimers across tion by enzymes belonging to the nitrilase family (Pace and the one-start helical groove. -

Sulfite Dehydrogenases in Organotrophic Bacteria : Enzymes
Sulfite dehydrogenases in organotrophic bacteria: enzymes, genes and regulation. Dissertation zur Erlangung des akademischen Grades des Doktors der Naturwissenschaften (Dr. rer. nat.) an der Universität Konstanz Fachbereich Biologie vorgelegt von Sabine Lehmann Tag der mündlichen Prüfung: 10. April 2013 1. Referent: Prof. Dr. Bernhard Schink 2. Referent: Prof. Dr. Andrew W. B. Johnston So eine Arbeit wird eigentlich nie fertig, man muss sie für fertig erklären, wenn man nach Zeit und Umständen das möglichste getan hat. (Johann Wolfgang von Goethe, Italienische Reise, 1787) DANKSAGUNG An dieser Stelle möchte ich mich herzlich bei folgenden Personen bedanken: . Prof. Dr. Alasdair M. Cook (Universität Konstanz, Deutschland), der mir dieses Thema und seine Laboratorien zur Verfügung stellte, . Prof. Dr. Bernhard Schink (Universität Konstanz, Deutschland), für seine spontane und engagierte Übernahme der Betreuung, . Prof. Dr. Andrew W. B. Johnston (University of East Anglia, UK), für seine herzliche und bereitwillige Aufnahme in seiner Arbeitsgruppe, seiner engagierten Unter- stützung, sowie für die Übernahme des Koreferates, . Prof. Dr. Frithjof C. Küpper (University of Aberdeen, UK), für seine große Hilfsbereitschaft bei der vorliegenden Arbeit und geplanter Manuskripte, als auch für die mentale Unterstützung während der letzten Jahre! Desweiteren möchte ich herzlichst Dr. David Schleheck für die Übernahme des Koreferates der mündlichen Prüfung sowie Prof. Dr. Alexander Bürkle, für die Übernahme des Prüfungsvorsitzes sowie für seine vielen hilfreichen Ratschläge danken! Ein herzliches Dankeschön geht an alle beteiligten Arbeitsgruppen der Universität Konstanz, der UEA und des SAMS, ganz besonders möchte ich dabei folgenden Personen danken: . Dr. David Schleheck und Karin Denger, für die kritische Durchsicht dieser Arbeit, der durch und durch sehr engagierten Hilfsbereitschaft bei Problemen, den zahlreichen wissenschaftlichen Diskussionen und für die aufbauenden Worte, . -

Cyanide-Degrading Enzymes for Bioremediation A
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Texas A&M University CYANIDE-DEGRADING ENZYMES FOR BIOREMEDIATION A Thesis by LACY JAMEL BASILE Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2008 Major Subject: Microbiology CYANIDE-DEGRADING ENZYMES FOR BIOREMEDIATION A Thesis by LACY JAMEL BASILE Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Approved by: Chair of Committee, Michael Benedik Committee Members, Susan Golden James Hu Wayne Versaw Head of Department, Vincent Cassone August 2008 Major Subject: Microbiology iii ABSTRACT Cyanide-Degrading Enzymes for Bioremediation. (August 2008) Lacy Jamel Basile, B.S., Texas A&M University Chair of Advisory Committee: Dr. Michael Benedik Cyanide-containing waste is an increasingly prevalent problem in today’s society. There are many applications that utilize cyanide, such as gold mining and electroplating, and these processes produce cyanide waste with varying conditions. Remediation of this waste is necessary to prevent contamination of soils and water. While there are a variety of processes being used, bioremediation is potentially a more cost effective alternative. A variety of fungal species are known to degrade cyanide through the action of cyanide hydratases, a specialized subset of nitrilases which hydrolyze cyanide to formamide. Here I report on previously unknown and uncharacterized nitrilases from Neurospora crassa, Gibberella zeae, and Aspergillus nidulans. Recombinant forms of four cyanide hydratases from N. -

Restriction Endonucleases
Molecular Biology Problem Solver: A Laboratory Guide. Edited by Alan S. Gerstein Copyright © 2001 by Wiley-Liss, Inc. ISBNs: 0-471-37972-7 (Paper); 0-471-22390-5 (Electronic) 9 Restriction Endonucleases Derek Robinson, Paul R. Walsh, and Joseph A. Bonventre Background Information . 226 Which Restriction Enzymes Are Commercially Available? . 226 Why Are Some Enzymes More Expensive Than Others? . 227 What Can You Do to Reduce the Cost of Working with Restriction Enzymes? . 228 If You Could Select among Several Restriction Enzymes for Your Application, What Criteria Should You Consider to Make the Most Appropriate Choice? . 229 What Are the General Properties of Restriction Endonucleases? . 232 What Insight Is Provided by a Restriction Enzyme’s Quality Control Data? . 233 How Stable Are Restriction Enzymes? . 236 How Stable Are Diluted Restriction Enzymes? . 236 Simple Digests . 236 How Should You Set up a Simple Restriction Digest? . 236 Is It Wise to Modify the Suggested Reaction Conditions? . 237 Complex Restriction Digestions . 239 How Can a Substrate Affect the Restriction Digest? . 239 Should You Alter the Reaction Volume and DNA Concentration? . 241 Double Digests: Simultaneous or Sequential? . 242 225 Genomic Digests . 244 When Preparing Genomic DNA for Southern Blotting, How Can You Determine If Complete Digestion Has Been Obtained? . 244 What Are Your Options If You Must Create Additional Rare or Unique Restriction Sites? . 247 Troubleshooting . 255 What Can Cause a Simple Restriction Digest to Fail? . 255 The Volume of Enzyme in the Vial Appears Very Low. Did Leakage Occur during Shipment? . 259 The Enzyme Shipment Sat on the Shipping Dock for Two Days. -

Structural and Biochemical Characterizations of Three Potential Drug Targets from Pathogens
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 2020 Structural and Biochemical Characterizations of Three Potential Drug Targets from Pathogens LU LU ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 ISBN 978-91-513-1148-7 UPPSALA urn:nbn:se:uu:diva-435815 2021 Dissertation presented at Uppsala University to be publicly examined in Room A1:111a, BMC, Husargatan 3, Uppsala, Friday, 16 April 2021 at 13:15 for the degree of Doctor of Philosophy. The examination will be conducted in English. Faculty examiner: Christian Cambillau. Abstract Lu, L. 2021. Structural and Biochemical Characterizations of Three Potential Drug Targets from Pathogens. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 2020. 91 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-513-1148-7. As antibiotic resistance of various pathogens emerged globally, the need for new effective drugs with novel modes of action became urgent. In this thesis, we focus on infectious diseases, e.g. tuberculosis, malaria, and nosocomial infections, and the corresponding causative pathogens, Mycobacterium tuberculosis, Plasmodium falciparum, and the Gram-negative ESKAPE pathogens that underlie so many healthcare-acquired diseases. Following the same- target-other-pathogen (STOP) strategy, we attempted to comprehensively explore the properties of three promising drug targets. Signal peptidase I (SPase I), existing both in Gram-negative and Gram-positive bacteria, as well as in parasites, is vital for cell viability, due to its critical role in signal peptide cleavage, thus, protein maturation, and secreted protein transport. Three factors, comprising essentiality, a unique mode of action, and easy accessibility, make it an attractive drug target. -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -

SUPPLEMENTARY INFORMATION in Silico Signature Prediction
SUPPLEMENTARY INFORMATION In Silico Signature Prediction Modeling in Cytolethal Distending Toxin-Producing Escherichia coli Strains Maryam Javadi, Mana Oloomi*, Saeid Bouzari Department of Molecular Biology, Pasteur Institute of Iran, Tehran 13164, Iran http://www.genominfo.org/src/sm/gni-15-69-s001.pdf Supplementary Table 6. Aalphabetic abbreviation and description of putative conserved domains Alphabetic Abbreviation Description 17 Large terminase protein 2_A_01_02 Multidrug resistance protein 2A0115 Benzoate transport; [Transport and binding proteins, Carbohydrates, organic alcohols] 52 DNA topisomerase II medium subunit; Provisional AAA_13 AAA domain; This family of domains contain a P-loop motif AAA_15 AAA ATPase domain; This family of domains contain a P-loop motif AAA_21 AAA domain AAA_23 AAA domain ABC_RecF ATP-binding cassette domain of RecF; RecF is a recombinational DNA repair ATPase ABC_SMC_barmotin ATP-binding cassette domain of barmotin, a member of the SMC protein family AcCoA-C-Actrans Acetyl-CoA acetyltransferases AHBA_syn 3-Amino-5-hydroxybenzoic acid synthase family (AHBA_syn) AidA Type V secretory pathway, adhesin AidA [Cell envelope biogenesis] Ail_Lom Enterobacterial Ail/Lom protein; This family consists of several bacterial and phage Ail_Lom proteins AIP3 Actin interacting protein 3; Aip3p/Bud6p is a regulator of cell and cytoskeletal polarity Aldose_epim_Ec_YphB Aldose 1-epimerase, similar to Escherichia coli YphB AlpA Predicted transcriptional regulator [Transcription] AntA AntA/AntB antirepressor AraC AraC-type -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection.