De Novo Transcriptome Assembly from the Gonads of a Scleractinian Coral
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SEDIMENTARY FRAMEWORK of Lmainland FRINGING REEF DEVELOPMENT, CAPE TRIBULATION AREA
GREAT BARRIER REEF MARINE PARK AUTHORITY TECHNICAL MEMORANDUM GBRMPA-TM-14 SEDIMENTARY FRAMEWORK OF lMAINLAND FRINGING REEF DEVELOPMENT, CAPE TRIBULATION AREA D.P. JOHNSON and RM.CARTER Department of Geology James Cook University of North Queensland Townsville, Q 4811, Australia DATE November, 1987 SUMMARY Mainland fringing reefs with a diverse coral fauna have developed in the Cape Tribulation area primarily upon coastal sedi- ment bodies such as beach shoals and creek mouth bars. Growth on steep rocky headlands is minor. The reefs have exten- sive sandy beaches to landward, and an irregular outer margin. Typically there is a raised platform of dead nef along the outer edge of the reef, and dead coral columns lie buried under the reef flat. Live coral growth is restricted to the outer reef slope. Seaward of the reefs is a narrow wedge of muddy, terrigenous sediment, which thins offshore. Beach, reef and inner shelf sediments all contain 50% terrigenous material, indicating the reefs have always grown under conditions of heavy terrigenous influx. The relatively shallow lower limit of coral growth (ca 6m below ADD) is typical of reef growth in turbid waters, where decreased light levels inhibit coral growth. Radiocarbon dating of material from surveyed sites confirms the age of the fossil coral columns as 33304110 ybp, indicating that they grew during the late postglacial sea-level high (ca 5500-6500 ybp). The former thriving reef-flat was killed by a post-5500 ybp sea-level fall of ca 1 m. Although this study has not assessed the community structure of the fringing reefs, nor whether changes are presently occur- ring, it is clear the corals present today on the fore-reef slope have always lived under heavy terrigenous influence, and that the fossil reef-flat can be explained as due to the mid-Holocene fall in sea-level. -

Response of Fluorescence Morphs of the Mesophotic Coral Euphyllia Paradivisa to Ultra-Violet Radiation
www.nature.com/scientificreports OPEN Response of fuorescence morphs of the mesophotic coral Euphyllia paradivisa to ultra-violet radiation Received: 23 August 2018 Or Ben-Zvi 1,2, Gal Eyal 1,2,3 & Yossi Loya 1 Accepted: 15 March 2019 Euphyllia paradivisa is a strictly mesophotic coral in the reefs of Eilat that displays a striking color Published: xx xx xxxx polymorphism, attributed to fuorescent proteins (FPs). FPs, which are used as visual markers in biomedical research, have been suggested to serve as photoprotectors or as facilitators of photosynthesis in corals due to their ability to transform light. Solar radiation that penetrates the sea includes, among others, both vital photosynthetic active radiation (PAR) and ultra-violet radiation (UVR). Both types, at high intensities, are known to have negative efects on corals, ranging from cellular damage to changes in community structure. In the present study, fuorescence morphs of E. paradivisa were used to investigate UVR response in a mesophotic organism and to examine the phenomenon of fuorescence polymorphism. E. paradivisa, although able to survive in high-light environments, displayed several physiological and behavioral responses that indicated severe light and UVR stress. We suggest that high PAR and UVR are potential drivers behind the absence of this coral from shallow reefs. Moreover, we found no signifcant diferences between the diferent fuorescence morphs’ responses and no evidence of either photoprotection or photosynthesis enhancement. We therefore suggest that FPs in mesophotic corals might have a diferent biological role than that previously hypothesized for shallow corals. Te solar radiation that reaches the earth’s surface includes, among others, ultra-violet radiation (UVR; 280– 400 nm) and photosynthetically active radiation (PAR; 400–700 nm). -

Resurrecting a Subgenus to Genus: Molecular Phylogeny of Euphyllia and Fimbriaphyllia (Order Scleractinia; Family Euphylliidae; Clade V)
Resurrecting a subgenus to genus: molecular phylogeny of Euphyllia and Fimbriaphyllia (order Scleractinia; family Euphylliidae; clade V) Katrina S. Luzon1,2,3,*, Mei-Fang Lin4,5,6,*, Ma. Carmen A. Ablan Lagman1,7, Wilfredo Roehl Y. Licuanan1,2,3 and Chaolun Allen Chen4,8,9,* 1 Biology Department, De La Salle University, Manila, Philippines 2 Shields Ocean Research (SHORE) Center, De La Salle University, Manila, Philippines 3 The Marine Science Institute, University of the Philippines, Quezon City, Philippines 4 Biodiversity Research Center, Academia Sinica, Taipei, Taiwan 5 Department of Molecular and Cell Biology, James Cook University, Townsville, Australia 6 Evolutionary Neurobiology Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan 7 Center for Natural Sciences and Environmental Research (CENSER), De La Salle University, Manila, Philippines 8 Taiwan International Graduate Program-Biodiversity, Academia Sinica, Taipei, Taiwan 9 Institute of Oceanography, National Taiwan University, Taipei, Taiwan * These authors contributed equally to this work. ABSTRACT Background. The corallum is crucial in building coral reefs and in diagnosing systematic relationships in the order Scleractinia. However, molecular phylogenetic analyses revealed a paraphyly in a majority of traditional families and genera among Scleractinia showing that other biological attributes of the coral, such as polyp morphology and reproductive traits, are underutilized. Among scleractinian genera, the Euphyllia, with nine nominal species in the Indo-Pacific region, is one of the groups Submitted 30 May 2017 that await phylogenetic resolution. Multiple genetic markers were used to construct Accepted 31 October 2017 Published 4 December 2017 the phylogeny of six Euphyllia species, namely E. ancora, E. divisa, E. -

Scleractinia Fauna of Taiwan I
Scleractinia Fauna of Taiwan I. The Complex Group 台灣石珊瑚誌 I. 複雜類群 Chang-feng Dai and Sharon Horng Institute of Oceanography, National Taiwan University Published by National Taiwan University, No.1, Sec. 4, Roosevelt Rd., Taipei, Taiwan Table of Contents Scleractinia Fauna of Taiwan ................................................................................................1 General Introduction ........................................................................................................1 Historical Review .............................................................................................................1 Basics for Coral Taxonomy ..............................................................................................4 Taxonomic Framework and Phylogeny ........................................................................... 9 Family Acroporidae ............................................................................................................ 15 Montipora ...................................................................................................................... 17 Acropora ........................................................................................................................ 47 Anacropora .................................................................................................................... 95 Isopora ...........................................................................................................................96 Astreopora ......................................................................................................................99 -

Jan 2021 London Zoo Stocklist.Pdf (596.63
ZSL London Zoo - January 2021 stocklist Status at 01.01.2021 m f unk Invertebrata Aurelia aurita * Moon jellyfish 0 0 150 Pachyclavularia violacea * Purple star coral 0 0 1 Tubipora musica * Organ-pipe coral 0 0 2 Pinnigorgia sp. * Sea fan 0 0 20 Sarcophyton sp. * Leathery soft coral 0 0 5 Sinularia sp. * Leathery soft coral 0 0 18 Sinularia dura * Cabbage leather coral 0 0 4 Sinularia polydactyla * Many-fingered leather coral 0 0 3 Xenia sp. * Yellow star coral 0 0 1 Heliopora coerulea * Blue coral 0 0 12 Entacmaea quadricolor Bladdertipped anemone 0 0 1 Epicystis sp. * Speckled anemone 0 0 1 Phymanthus crucifer * Red beaded anemone 0 0 11 Heteractis sp. * Elegant armed anemone 0 0 1 Stichodactyla tapetum Mini carpet anemone 0 0 1 Discosoma sp. * Umbrella false coral 0 0 21 Rhodactis sp. * Mushroom coral 0 0 8 Ricordea sp. * Emerald false coral 0 0 19 Acropora sp. * Staghorn coral 0 0 115 Acropora humilis * Staghorn coral 0 0 1 Acropora yongei * Staghorn coral 0 0 2 Montipora sp. * Montipora coral 0 0 5 Montipora capricornis * Coral 0 0 5 Montipora confusa * Encrusting coral 0 0 22 Montipora danae * Coral 0 0 23 Montipora digitata * Finger coral 0 0 6 Montipora foliosa * Hard coral 0 0 10 Montipora hodgsoni * Coral 0 0 2 Pocillopora sp. * Cauliflower coral 0 0 27 Seriatopora hystrix * Bird nest coral 0 0 8 Stylophora sp. * Cauliflower coral 0 0 1 Stylophora pistillata * Pink cauliflower coral 0 0 23 Catalaphyllia jardinei * Elegance coral 0 0 4 Euphyllia ancora * Crescent coral 0 0 4 Euphyllia glabrescens * Joker's cap coral 0 0 2 Euphyllia paradivisa * Branching frog spawn 0 0 3 Euphyllia paraancora * Branching hammer coral 0 0 3 Euphyllia yaeyamaensis * Crescent coral 0 0 4 Plerogyra sinuosa * Bubble coral 0 0 1 Duncanopsammia axifuga + Coral 0 0 2 Tubastraea sp. -
Jan 2021 ZSL Stocklist.Pdf (699.26
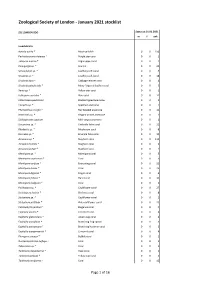
Zoological Society of London - January 2021 stocklist ZSL LONDON ZOO Status at 01.01.2021 m f unk Invertebrata Aurelia aurita * Moon jellyfish 0 0 150 Pachyclavularia violacea * Purple star coral 0 0 1 Tubipora musica * Organ-pipe coral 0 0 2 Pinnigorgia sp. * Sea fan 0 0 20 Sarcophyton sp. * Leathery soft coral 0 0 5 Sinularia sp. * Leathery soft coral 0 0 18 Sinularia dura * Cabbage leather coral 0 0 4 Sinularia polydactyla * Many-fingered leather coral 0 0 3 Xenia sp. * Yellow star coral 0 0 1 Heliopora coerulea * Blue coral 0 0 12 Entacmaea quadricolor Bladdertipped anemone 0 0 1 Epicystis sp. * Speckled anemone 0 0 1 Phymanthus crucifer * Red beaded anemone 0 0 11 Heteractis sp. * Elegant armed anemone 0 0 1 Stichodactyla tapetum Mini carpet anemone 0 0 1 Discosoma sp. * Umbrella false coral 0 0 21 Rhodactis sp. * Mushroom coral 0 0 8 Ricordea sp. * Emerald false coral 0 0 19 Acropora sp. * Staghorn coral 0 0 115 Acropora humilis * Staghorn coral 0 0 1 Acropora yongei * Staghorn coral 0 0 2 Montipora sp. * Montipora coral 0 0 5 Montipora capricornis * Coral 0 0 5 Montipora confusa * Encrusting coral 0 0 22 Montipora danae * Coral 0 0 23 Montipora digitata * Finger coral 0 0 6 Montipora foliosa * Hard coral 0 0 10 Montipora hodgsoni * Coral 0 0 2 Pocillopora sp. * Cauliflower coral 0 0 27 Seriatopora hystrix * Bird nest coral 0 0 8 Stylophora sp. * Cauliflower coral 0 0 1 Stylophora pistillata * Pink cauliflower coral 0 0 23 Catalaphyllia jardinei * Elegance coral 0 0 4 Euphyllia ancora * Crescent coral 0 0 4 Euphyllia glabrescens * Joker's cap coral 0 0 2 Euphyllia paradivisa * Branching frog spawn 0 0 3 Euphyllia paraancora * Branching hammer coral 0 0 3 Euphyllia yaeyamaensis * Crescent coral 0 0 4 Plerogyra sinuosa * Bubble coral 0 0 1 Duncanopsammia axifuga + Coral 0 0 2 Tubastraea sp. -

Molecular Approaches Underlying the Oogenic Cycle of the Scleractinian
www.nature.com/scientificreports OPEN Molecular approaches underlying the oogenic cycle of the scleractinian coral, Acropora tenuis Ee Suan Tan1, Ryotaro Izumi1, Yuki Takeuchi 2,3, Naoko Isomura4 & Akihiro Takemura2 ✉ This study aimed to elucidate the physiological processes of oogenesis in Acropora tenuis. Genes/ proteins related to oogenesis were investigated: Vasa, a germ cell marker, vitellogenin (VG), a major yolk protein precursor, and its receptor (LDLR). Coral branches were collected monthly from coral reefs around Sesoko Island (Okinawa, Japan) for histological observation by in situ hybridisation (ISH) of the Vasa (AtVasa) and Low Density Lipoprotein Receptor (AtLDLR) genes and immunohistochemistry (IHC) of AtVasa and AtVG. AtVasa immunoreactivity was detected in germline cells and ooplasm, whereas AtVG immunoreactivity was detected in ooplasm and putative ovarian tissues. AtVasa was localised in germline cells located in the retractor muscles of the mesentery, whereas AtLDLR was localised in the putative ovarian and mesentery tissues. AtLDLR was detected in coral tissues during the vitellogenic phase, whereas AtVG immunoreactivity was found in primary oocytes. Germline cells expressing AtVasa are present throughout the year. In conclusion, Vasa has physiological and molecular roles throughout the oogenic cycle, as it determines gonadal germline cells and ensures normal oocyte development, whereas the roles of VG and LDLR are limited to the vitellogenic stages because they act in coordination with lipoprotein transport, vitellogenin synthesis, and yolk incorporation into oocytes. Approximately 70% of scleractinian corals are hermaphroditic broadcast spawners and have both male and female gonads developing within the polyp of the same colony1. Tey engage in a multispecifc spawning event around the designated moon phase once a year2–4. -

Recent Advances in Lagoon-Based Farming Practices for Eight Species of Commercially Valuable Hard and Soft Corals - a Technical Report
CTSA Publication No. 147 Recent Advances in Lagoon-based Farming Practices for Eight Species of Commercially Valuable Hard and Soft Corals - A Technical Report Simon Ellis Eileen Ellis Recent Advances in Lagoon-based Farming Practices for Eight Species of Commercially Valuable Hard and Soft Corals - A Technical Report Simon Ellis Eileen Ellis Center for Tropical and Subtropical Aquaculture Publication No. 147 March 2002 Table of Contents Table of Contents Acknowledgments .......................................................................... 1 Foreword .......................................................................................... 2 Introduction...................................................................................... 3 About this publication ................................................................... 3 The world market for live corals ................................................... 4 Coral species used in the study ................................................... 5 Site selection ................................................................................... 9 Test sites ...................................................................................... 9 Advances in farm structure ......................................................... 11 A modular approach ................................................................... 11 Use of cages .............................................................................. 14 Broodstock ................................................................................... -

Seventy-Four Universal Primers for Characterizing the Complete Mitochondrial Genomes of Scleractinian Corals (Cnidaria; Anthozoa) Mei-Fang Lin1,5, Katrina S
Zoological Studies 50(4): 513-524 (2011) Seventy-four Universal Primers for Characterizing the Complete Mitochondrial Genomes of Scleractinian Corals (Cnidaria; Anthozoa) Mei-Fang Lin1,5, Katrina S. Luzon2, Wilfredo Y. Licuanan3, Maria Carmen Ablan-Lagman4, and Chaolun A. Chen1,5,6,7,* 1Biodiversity Research Center, Academia Sinica, Nankang, Taipei 115, Taiwan 2The Marine Science Institute, Univ. of the Philippines, Quezon City 1101, the Philippines 3Biology Department and Brother Alfred Shields Marine Station, De La Salle Univ., Manila 1004, the Philippines 4Biology Department, De La Salle Univ., Manila,1004, the Philippines 5Institute of Oceanography, National Taiwan Univ., Taipei 106, Taiwan 6Institute of Life Science, National Taitung Univ., Taitung 904, Taiwan 7ARC Centre of Excellence for Coral Reef Studies, James Cook Univ., Townsville 4810, Australia (Accepted January 28, 2011) Mei-Fang Lin, Katrina S. Luzon, Wilfredo Y. Licuanan, Maria Carmen Ablan-Lagman, and Chaolun A. Chen (2011) Seventy-four universal primers for characterizing the complete mitochondrial genomes of scleractinian corals (Cnidaria; Anthozoa). Zoological Studies 50(4): 513-524. Use of universal primers designed from a public DNA database can accelerate characterization of mitochondrial (mt) genomes for targeted taxa by polymerase chain reaction (PCR) amplification and direct DNA sequencing. This approach can obtain large amounts of mt information for phylogenetic inferences at lower costs and in less time. In this study, 88 primers were designed from 13 published scleractinian mt genomes, and these were tested on Euphyllia ancora, Galaxea fascicularis, Fungiacyathus stephanus, Porites okinawensis, Goniopora columna, Tubastraea coccinea, Pavona venosa, Oulastrea crispata, and Polycyathus sp., representing 7 families of complex and robust corals. -

Coral Reef Species List
Coral Reef Gallery The Philippine Coral Reef Tank focuses on the most diverse and fragile of marine ecosystems. From the main exhibit floor, visitors look down on a shallow, sandy lagoon—a calm, protected area inhabited by sharks, rays, and colorful fishes. Where the lagoon drops off to the deep reef, hundreds of bright fishes visible near the surface lure the visitor to view the spectacle one floor below. There, dramatic underwater views of the deep reef invite contemplation. Featuring 1,000 square feet of living coral and some 4,000 fish of 100 or more species, this 212,000‐gallon exhibit is, at 25 feet, the deepest and one of the largest displays of a living coral reef in the world. Curiosity leads to exploration of several smaller galleries along the perimeter of the exhibit that highlight the unique adaptations and complex interactions of reef organisms. Acanthastrea echinata Acanthurus achilles Acanthurus blochii Acanthurus coeruleus Blue tang Acanthurus dussumieri Acanthurus japonicus Acanthurus lineatus Acanthurus mata Acanthurus nigricans Acanthurus nigrofuscus Acanthurus nigroris Acanthurus olivaceus Acanthurus pyroferus Acanthurus triostegus Acanthurus xanthopterus Acropora formosa Acropora gemmifera Acropora micropthalma Acropora millepora Acropora sp. Staghorn Coral Acropora youngei Aeoliscus strigatus Shrimpfish Alcyonium sp. Alpheus randalli Randall’s Partner Shrimp Alveopora sp. Ambligobius hectori Hector’s Goby Ambligobius rainfordi Rainford’s Goby Amblycirrhitus pinos Redspotted hawkfish Amblyeleotris randalli Randall’s -

Manado Corals (866) 874-7639 (855) 225-8086
American Ingenuity Tranship www.livestockusa.org Manado Corals (866) 874-7639 (855) 225-8086 Assorted only with some high-end in every box Tranship - F.O.B. Manado Sunday to LAX - Monday to You Animal cost plus landing costs Order cut-off is Thursday See landing costs below Arrives to LAX on May 2, 2021 Please note: List is directly from Manado, our apologies for any typos! Box # Code Common Name Binomial - scientific name Qty in Box Price ea. 14 70504.4 Catalaphyllia jardinei Elegance Coral Purple Tip S 1 $37.50 71304.3 Euphyllia Cristata Branch frogtip Green Met M 1 $57.00 71403.3 Euphyllia ancora Wall Hammer Orange Metalic M 1 $74.25 71406.1 Euphyllia ancora Wall hammer Green SUPER 1 $111.75 71705 Favites sp. Pinneapple Met Green 1 $24.75 71903.1 Galaxea sp. Crystal Coral Green Met 1 $24.75 72104 Goniopora sp. Red Yellow Eye Encrusting Goniopora 1 $87.00 72110 Goniopora sp. Sunflower Green Metallic 1 $11.25 72204.3 Heliofungia actinoformis Long Tentacle Plate Green Met M 1 $37.50 73203.3 Plerogyra sinuosa Bubble Cat Eyes Green M 1 $45.00 73713.3 Wellsophyllia radiata Flat Brain Bicolor Met M 1 $87.00 74001 Tubipora musica Organ Pipe Green Met 1 $24.75 80202 Pachyclavularia sp Star Polyps Met Green 1 $21.75 80403 Palythoa Long stalk polyp Met color 1 $24.75 80405 Palythoa Long stalk polyp Blue SUPER 1 $99.00 80506 Parazoanthus spp. Sand polyp metallic color 1 $15.00 80603.1 Protopalythoa sp. Pipe Button Polyp Red Metalic Makasar 1 $62.25 80702 Actinodiscus sp. -

Hyperspectral Imaging As a Tool for Assessing Coral Health Utilising Natural Fluorescence
Teague, J. , Willans, J., Allen, M., Scott, T., & Day, J. (2019). Hyperspectral imaging as a tool for assessing coral health utilising natural fluorescence. Journal of Spectral Imaging, 8, [a7]. https://doi.org/10.1255/jsi.2019.a7 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1255/jsi.2019.a7 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via IMPublications at https://www.impopen.com/jsi-abstract/I08_a7 . Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ J. Teague et al., J. Spectral Imaging 8, a7 (2019) 1 volume 1 / 2010 JsI ISSN 2040-4565 JOURNAL OF SPECTRALPeer Reviewed Paper Paper Presented at HSI 2018, October 2018, Coventry, UK IMAGING openaccess In thIs Issue: spectral preprocessing to compensate for packaging film / using neural nets to invert Hyperspectralthe PROSAIL canopy model imaging as a tool for assessing coral health utilising natural fluorescence Jonathan Teague,a,* Jack Willans,b Michael J. Allen,c Thomas B. Scottd and John C.C. Daye aInterface Analysis Centre (IAC), HH Wills Physics Laboratory, Tyndall Ave, Bristol BS8 1TL, UK. E-mail: [email protected], https://orcid.org/0000-0001-7817-6434 bSealife London Aquarium, County Hall, Westminster Bridge Rd, Lambeth, London SE1 7PB, UK.