That Had Torte I Una Altra Manian Literatura
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Toronto Research Chemicals Products for Innovative Research
My Account Checkout Register Cart Toronto Research Chemicals products for innovative research Search Products Cannabis Testing Products Screening Library Services Company Profile Featured Products Careers Support Contact Us Blog E889200 Etafedrine Hydrochloride (mixture of diastereomers) Catalogue number: E889200 Chemical name: Etafedrine Hydrochloride (mixture of diastereomers) Synonyms: Ethylephedrine Hydrochloride; α-[1-(Ethylmethylamino)ethyl]benzenemethanol Hydrochloride; α-[1-(Ethylmethylamino)ethyl]benzyl Alcohol Hydrochloride; Dit enate; Novedrine Hydrochloride; Impurity: CAS Number: 5591-29-7 Possible CAS #: NA Molecular form.: C₁₂H₂₀ClNO Appearance: White to Off-White Solid Melting Point: 108-110°C Mol. Weight: 229.75 Storage: Hygroscopic, -20˚C Freezer, Under Inert Atmosphere Solubility: Chloroform (Slightly), Methanol (Slightly) Stability: No Data Available Category: Amines, Aromatics, Neurochemicals, Pharmaceuticals, Intermediates & Fine Ch emicals Boiling Point: No Data Available Applications: Etafedrine is the N-ethylated analogue of Ephedrine (E575000). Etafedrine is a l ong-acting bronchodilator that acts as a selective β-adrenergic receptor agonist . Dangerous Goods Info: NA References: Lindmar R. et al.: Arzneim.-Forsch., 35, 602 (1985); Ueda, T. et al.: Pharm. Bull., 3, 465 (1955); Peer Review Product Citations: MSDS: E889200.pdf Keywords: Buy 5591-29-7 | Purchase 5591-29-7 | Order 5591-29-7 | 5591-29-7 supplier | 5591-29-7 manufacturer | 5591-29-7 distributor | 5591-29-7 cost Order Product Pack size Price(USD) Qty 5mg $110.00 Qty 25mg $350.00 Qty 100mg $1200.00 Pack size Price(USD) Qty 10mg $150.00 Qty 50mg $685.00 Qty Exact Weight Packaging ($75(USD) / Vial) InventoryInventory Status:Status: In Stock Add to Cart Have a question about this product? Click here. -

Meat Biotechnology – Applications in Pork Quality Muscle Targeted Growth Promotants – Mode of Action of Beta Agonists
Meat Biotechnology – Applications in Pork Quality Muscle Targeted Growth Promotants – Mode of Action of Beta Agonists Deana Hancock, Diane Moody, Dave Anderson Elanco Animal Health Reciprocal Meat Conference, June 19, 2006 Pharmacological Activity: Beta•Adrenergic Agonist What is a beta•adrenergic agonist? A beta•adrenergic agonist (b•agonist) binds to and activates beta•adrenergic receptors (b•AR) that are found on the surface of many different types of cells in the body. G A G M L V L V G P A A A L P D G Beta Adrenergic P G L E N S S A S A T A N D P V L R A Y Receptors L L P L C K P A S P S R C P A P R C A A D S E F E S D E Y G V R E P W S S E P L T H L S R F E Q Q N F V W G F A R A P T W C R A K D A E L W Y V R G V V I W A V L (1) M (3) W H (5) I N (7) F G T T S L A L L G A S L M A F V M F V S F F A D I N L P V V P V F V L Shaded AA's are V I L C L F S V L P L W Transmembrane L G L S F V T Y W G conserved across the L I M A V L V P C Y A Domains V L A L V E I S S G A S A D A I S I C L V F T N human b1, b2 and b3 T L M A N A L W V A G M F AR's (31%) V L C V T F I N V M S C V I P I I I V I V Y G A I A L L L I A (2) F L L (4) G R (6) T Y K N D R V K C A L R F A R T T G L L C Y R R K S Q C A R R P L A F A R L E Q P D F R K A L T Q A R A E A T Q R R I K P H T L L S A R R D G A H R R T Q R L V A G L V S C Circled AA's are S K P R A L S P P Q K D conserved across F R Y I R A R G D D D D R P D V V G A D K G G N P P S the b AR of S A P G A S T 1 C A E L P humans, pigs, R L L P R P P E A F G N W R and sheep (80%) L A A A A G C G A G G A A A R P A A T D P G P P P S S S L E S K A P D D E P A S V C R F R P P G A P P P S P S P S V A Mersmann, 1998; J. -

(19) United States (12) Patent Application Publication (10) Pub
US 20130289061A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0289061 A1 Bhide et al. (43) Pub. Date: Oct. 31, 2013 (54) METHODS AND COMPOSITIONS TO Publication Classi?cation PREVENT ADDICTION (51) Int. Cl. (71) Applicant: The General Hospital Corporation, A61K 31/485 (2006-01) Boston’ MA (Us) A61K 31/4458 (2006.01) (52) U.S. Cl. (72) Inventors: Pradeep G. Bhide; Peabody, MA (US); CPC """"" " A61K31/485 (201301); ‘4161223011? Jmm‘“ Zhu’ Ansm’ MA. (Us); USPC ......... .. 514/282; 514/317; 514/654; 514/618; Thomas J. Spencer; Carhsle; MA (US); 514/279 Joseph Biederman; Brookline; MA (Us) (57) ABSTRACT Disclosed herein is a method of reducing or preventing the development of aversion to a CNS stimulant in a subject (21) App1_ NO_; 13/924,815 comprising; administering a therapeutic amount of the neu rological stimulant and administering an antagonist of the kappa opioid receptor; to thereby reduce or prevent the devel - . opment of aversion to the CNS stimulant in the subject. Also (22) Flled' Jun‘ 24’ 2013 disclosed is a method of reducing or preventing the develop ment of addiction to a CNS stimulant in a subj ect; comprising; _ _ administering the CNS stimulant and administering a mu Related U‘s‘ Apphcatlon Data opioid receptor antagonist to thereby reduce or prevent the (63) Continuation of application NO 13/389,959, ?led on development of addiction to the CNS stimulant in the subject. Apt 27’ 2012’ ?led as application NO_ PCT/US2010/ Also disclosed are pharmaceutical compositions comprising 045486 on Aug' 13 2010' a central nervous system stimulant and an opioid receptor ’ antagonist. -

Ep 2560611 B1
(19) TZZ Z___T (11) EP 2 560 611 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: A61K 9/00 (2006.01) 03.01.2018 Bulletin 2018/01 (86) International application number: (21) Application number: 11719211.2 PCT/EP2011/056227 (22) Date of filing: 19.04.2011 (87) International publication number: WO 2011/131663 (27.10.2011 Gazette 2011/43) (54) "PROCESS FOR PROVIDING PARTICLES WITH REDUCED ELECTROSTATIC CHARGES" VERFAHREN ZUR BEREITSTELLUNG VON PARTIKELN MIT REDUZIERTEN ELEKTROSTATISCHEN LADUNGEN PROCÉDÉ DE PRÉPARATION DE PARTICULES AYANT DES CHARGES ÉLECTROSTATIQUES RÉDUITES (84) Designated Contracting States: • GUCHARDI ET AL: "Influence of fine lactose and AL AT BE BG CH CY CZ DE DK EE ES FI FR GB magnesium stearate on low dose dry powder GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO inhaler formulations", INTERNATIONAL PL PT RO RS SE SI SK SM TR JOURNAL OF PHARMACEUTICS, ELSEVIER BV, NL LNKD- DOI:10.1016/J.IJPHARM.2007.06.041, (30) Priority: 21.04.2010 EP 10160565 vol. 348, no. 1-2, 19 December 2007 (2007-12-19), pages 10-17, XP022393884, ISSN: 0378-5173 (43) Date of publication of application: • ELAJNAF A ET AL: "Electrostatic 27.02.2013 Bulletin 2013/09 characterisation of inhaled powders: Effect of contact surface and relative humidity", (73) Proprietor: Chiesi Farmaceutici S.p.A. EUROPEAN JOURNAL OF PHARMACEUTICAL 43100 Parma (IT) SCIENCES, ELSEVIER, AMSTERDAM, NL LNKD- DOI:10.1016/J.EJPS.2006.07.006, vol. 29, no. 5, 1 (72) Inventors: December 2006 (2006-12-01), pages 375-384, • COCCONI, Daniela XP025137181, ISSN: 0928-0987 [retrieved on I-43100 Parma (IT) 2006-12-01] • MUSA, Rossella • CHAN ET AL: "Dry powder aerosol drug I-43100 Parma (IT) delivery-Opportunities for colloid and surface scientists", COLLOIDS AND SURFACES. -

Customs Tariff - Schedule
CUSTOMS TARIFF - SCHEDULE 99 - i Chapter 99 SPECIAL CLASSIFICATION PROVISIONS - COMMERCIAL Notes. 1. The provisions of this Chapter are not subject to the rule of specificity in General Interpretative Rule 3 (a). 2. Goods which may be classified under the provisions of Chapter 99, if also eligible for classification under the provisions of Chapter 98, shall be classified in Chapter 98. 3. Goods may be classified under a tariff item in this Chapter and be entitled to the Most-Favoured-Nation Tariff or a preferential tariff rate of customs duty under this Chapter that applies to those goods according to the tariff treatment applicable to their country of origin only after classification under a tariff item in Chapters 1 to 97 has been determined and the conditions of any Chapter 99 provision and any applicable regulations or orders in relation thereto have been met. 4. The words and expressions used in this Chapter have the same meaning as in Chapters 1 to 97. Issued January 1, 2020 99 - 1 CUSTOMS TARIFF - SCHEDULE Tariff Unit of MFN Applicable SS Description of Goods Item Meas. Tariff Preferential Tariffs 9901.00.00 Articles and materials for use in the manufacture or repair of the Free CCCT, LDCT, GPT, UST, following to be employed in commercial fishing or the commercial MT, MUST, CIAT, CT, harvesting of marine plants: CRT, IT, NT, SLT, PT, COLT, JT, PAT, HNT, Artificial bait; KRT, CEUT, UAT, CPTPT: Free Carapace measures; Cordage, fishing lines (including marlines), rope and twine, of a circumference not exceeding 38 mm; Devices for keeping nets open; Fish hooks; Fishing nets and netting; Jiggers; Line floats; Lobster traps; Lures; Marker buoys of any material excluding wood; Net floats; Scallop drag nets; Spat collectors and collector holders; Swivels. -

(19) United States (12) Patent Application Publication (10) Pub
US 20050181041A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2005/0181041 A1 Goldman (43) Pub. Date: Aug. 18, 2005 (54) METHOD OF PREPARATION OF MIXED Related US. Application Data PHASE CO-CRYSTALS WITH ACTIVE AGENTS (60) Provisional application No. 60/528,232, ?led on Dec. 9, 2003. Provisional application No. 60/559,862, ?led (75) Inventor: David Goldman, Portland, CT (US) on Apr. 6, 2004. Correspondence Address: Publication Classi?cation LEYDIG VOIT & MAYER, LTD (51) Int. Cl.7 ....................... .. A61K 31/56; A61K 38/00; TWO PRUDENTIAL PLAZA, SUITE 4900 A61K 9/64 180 NORTH STETSON AVENUE (52) US. Cl. ............................ .. 424/456; 514/179; 514/2; CHICAGO, IL 60601-6780 (US) 514/221 (73) Assignee: MedCrystalForms, LLC, Hunt Valley, (57) ABSTRACT MD This invention pertains to a method of preparing mixed phase co-crystals of active agents With one or more materials (21) Appl. No.: 11/008,034 that alloWs the modi?cation of the active agent to a neW physical/crystal form With unique properties useful for the delivery of the active agent, as Well as compositions com (22) Filed: Dec. 9, 2004 prising the mixed phase co-crystals. Patent Application Publication Aug. 18, 2005 Sheet 1 0f 8 US 2005/0181041 A1 FIG. 1a 214.70°C z.m."m.n... 206.98°C n..0ao 142 OJ/g as:20m=3: -0.8 -1.0 40 90 1:10 2110 Temperture (°C) FIG. 1b 0.01 as:22“.Km: 217 095 24221.4 39Jmum/Q -0.8 35 155 255 255 Temperture (°C) Patent Application Publication Aug. -

Partial Agreement in the Social and Public Health Field
COUNCIL OF EUROPE COMMITTEE OF MINISTERS (PARTIAL AGREEMENT IN THE SOCIAL AND PUBLIC HEALTH FIELD) RESOLUTION AP (88) 2 ON THE CLASSIFICATION OF MEDICINES WHICH ARE OBTAINABLE ONLY ON MEDICAL PRESCRIPTION (Adopted by the Committee of Ministers on 22 September 1988 at the 419th meeting of the Ministers' Deputies, and superseding Resolution AP (82) 2) AND APPENDIX I Alphabetical list of medicines adopted by the Public Health Committee (Partial Agreement) updated to 1 July 1988 APPENDIX II Pharmaco-therapeutic classification of medicines appearing in the alphabetical list in Appendix I updated to 1 July 1988 RESOLUTION AP (88) 2 ON THE CLASSIFICATION OF MEDICINES WHICH ARE OBTAINABLE ONLY ON MEDICAL PRESCRIPTION (superseding Resolution AP (82) 2) (Adopted by the Committee of Ministers on 22 September 1988 at the 419th meeting of the Ministers' Deputies) The Representatives on the Committee of Ministers of Belgium, France, the Federal Republic of Germany, Italy, Luxembourg, the Netherlands and the United Kingdom of Great Britain and Northern Ireland, these states being parties to the Partial Agreement in the social and public health field, and the Representatives of Austria, Denmark, Ireland, Spain and Switzerland, states which have participated in the public health activities carried out within the above-mentioned Partial Agreement since 1 October 1974, 2 April 1968, 23 September 1969, 21 April 1988 and 5 May 1964, respectively, Considering that the aim of the Council of Europe is to achieve greater unity between its members and that this -
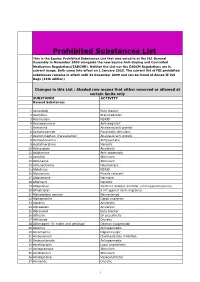
Prohibited Substances List
Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). Neither the List nor the EADCM Regulations are in current usage. Both come into effect on 1 January 2010. The current list of FEI prohibited substances remains in effect until 31 December 2009 and can be found at Annex II Vet Regs (11th edition) Changes in this List : Shaded row means that either removed or allowed at certain limits only SUBSTANCE ACTIVITY Banned Substances 1 Acebutolol Beta blocker 2 Acefylline Bronchodilator 3 Acemetacin NSAID 4 Acenocoumarol Anticoagulant 5 Acetanilid Analgesic/anti-pyretic 6 Acetohexamide Pancreatic stimulant 7 Acetominophen (Paracetamol) Analgesic/anti-pyretic 8 Acetophenazine Antipsychotic 9 Acetylmorphine Narcotic 10 Adinazolam Anxiolytic 11 Adiphenine Anti-spasmodic 12 Adrafinil Stimulant 13 Adrenaline Stimulant 14 Adrenochrome Haemostatic 15 Alclofenac NSAID 16 Alcuronium Muscle relaxant 17 Aldosterone Hormone 18 Alfentanil Narcotic 19 Allopurinol Xanthine oxidase inhibitor (anti-hyperuricaemia) 20 Almotriptan 5 HT agonist (anti-migraine) 21 Alphadolone acetate Neurosteriod 22 Alphaprodine Opiod analgesic 23 Alpidem Anxiolytic 24 Alprazolam Anxiolytic 25 Alprenolol Beta blocker 26 Althesin IV anaesthetic 27 Althiazide Diuretic 28 Altrenogest (in males and gelidngs) Oestrus suppression 29 Alverine Antispasmodic 30 Amantadine Dopaminergic 31 Ambenonium Cholinesterase inhibition 32 Ambucetamide Antispasmodic 33 Amethocaine Local anaesthetic 34 Amfepramone Stimulant 35 Amfetaminil Stimulant 36 Amidephrine Vasoconstrictor 37 Amiloride Diuretic 1 Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). -

Stability Studies of Amphetamine and Ephedrine Derivatives in Urine C
Journal of Chromatography B, 843 (2006) 84–93 Stability studies of amphetamine and ephedrine derivatives in urine C. Jimenez´ a,b, R. de la Torre a,b, M. Ventura a,c, J. Segura a,b, R. Ventura a,b,∗ a Unitat de Farmacologia, Institut Municipal d’Investigaci´oM`edica, Barcelona, Spain b CEXS, Universitat Pompeu Fabra, Barcelona, Spain c UDIMAS, Universitat Aut´onoma de Barcelona, Barcelona, Spain Received 3 March 2006; accepted 23 May 2006 Available online 23 June 2006 Abstract Knowledge of the stability of drugs in biological specimens is a critical consideration for the interpretation of analytical results. Identification of proper storage conditions has been a matter of concern for most toxicology laboratories (both clinical and forensic), and the stability of drugs of abuse has been extensively studied. This concern should be extended to other areas of analytical chemistry like antidoping control. In this work, the stability of ephedrine derivatives (ephedrine, norephedrine, methylephedrine, pseudoephedrine, and norpseudoephedrine), and amphetamine derivatives (amphetamine, methamphetamine, 3,4-methylenedioxyamphetamine (MDA), and 3,4-methylenedioxymethamphetamine (MDMA)) in urine has been studied. Spiked urine samples were prepared for stability testing. Urine samples were quantified by GC/NPD or GC/MS. The homogeneity of each batch of sample was verified before starting the stability study. The stability of analytes was evaluated in sterilized and non-sterilized urine samples at different storage conditions. For long-term stability testing, analyte concentration in urine stored at 4 ◦C and −20 ◦C was determined at different time intervals for 24 months for sterile urine samples, and for 6 months for non-sterile samples. -

Clenbuterol Elisa Kit Instructions Product #101219 & 101216 Forensic Use Only
Neogen Corporation 944 Nandino Blvd., Lexington KY 40511 USA 800/477-8201 USA/Canada | 859/254-1221 Fax: 859/255-5532 | E-mail: [email protected] | Web: www.neogen.com/Toxicology CLENBUTEROL ELISA KIT INSTRUCTIONS PRODUCT #101219 & 101216 FORENSIC USE ONLY INTENDED USE: For the determination of trace quantities of Clenbuterol and/or other metabolites in human urine, blood, oral fluid. DESCRIPTION Neogen Corporation’s Clenbuterol ELISA (Enzyme-Linked ImmunoSorbent Assay) test kit is a qualitative one-step kit designed for use as a screening device for the detection of drugs and/or their metabolites. The kit was designed for screening purposes and is intended for forensic use only. It is recommended that all suspect samples be confirmed by a quantitative method such as gas chromatography/mass spectrometry (GC/MS). ASSAY PRINCIPLES Neogen Corporation’s test kit operates on the basis of competition between the drug or its metabolite in the sample and the drug- enzyme conjugate for a limited number of antibody binding sites. First, the sample or control is added to the microplate. Next, the diluted drug-enzyme conjugate is added and the mixture is incubated at room temperature. During this incubation, the drug in the sample or the drug-enzyme conjugate binds to antibody immobilized in the microplate wells. After incubation, the plate is washed 3 times to remove any unbound sample or drug-enzyme conjugate. The presence of bound drug-enzyme conjugate is recognized by the addition of K-Blue® Substrate (TMB). After a 30 minute substrate incubation, the reaction is halted with the addition of Red Stop Solution. -

The Use of Stems in the Selection of International Nonproprietary Names (INN) for Pharmaceutical Substances
WHO/PSM/QSM/2006.3 The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances 2006 Programme on International Nonproprietary Names (INN) Quality Assurance and Safety: Medicines Medicines Policy and Standards The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 © World Health Organization 2006 All rights reserved. Publications of the World Health Organization can be obtained from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press, at the above address (fax: +41 22 791 4806; e-mail: [email protected]). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. -

CATO佳途科技- Cato Research Chemicals Inc
CATO 现货标准品产品清单 Cato Research Chemicals Inc. 广州佳途科技股份有限公司 CATO Research Chemicals Inc.专注于标准品研发、生产、服务及销 售,率先在中国建立亚洲技术中心实验室,并已通过ISO17034、 药物杂质标准品联系人:黄舒琦 ISO9001体系认证。CATO中国95%以上员工均为本科以上学历,具有高 电话:020-81215950 度专业的化工化学行业经验。 非药物标准品联系人:叶淑明 目前为止,CATO中国已服务超1万家客户,提供的标准品覆盖工业 电话:020-81960175 品、食品、农药残留、兽药残留、环境、药物杂质、天然提取物等领域, E-mail:[email protected] 同时深度支持定制合成。 “扫一扫” 地址:广州市荔湾区西增路63号自编E1-A101A 我们相信,CATO能给您更好的产品及服务! 关注CATO微信公众号 官网:http://www.cato-chem.com No. 货号 中文名 英文名 CAS号 药物杂质标准品 1 C3D-1016 2-氨基苯酚 2-Aminophenol 95-55-6 2 C3D-1331 门冬氨酸缩合物 (S)-2-((S)-3-Amino-3-carboxypropanamido)succinic acid 60079-22-3 3 C3D-1350 4-氯苯甲酸 4-Chlorobenzoic acid 74-11-3 4 C3D-1447 戊乙奎醚杂质4 Penehyclidine Impurity 4 5422-88-8 5 C3D-1481 氮卓斯汀EP杂质A Azelastine EP Impurity A 613-94-5 6 C3D-1593 氟西汀EP杂质A(盐酸托莫西汀相关化合物A) Fluoxetine EP Impurity A(Atomoxetine Related Compound A) 42142-52-9 7 C3D-1598 瑞巴派特 Rebamipide 90098-04-7 8 C3D-1636 4-甲基苯磺酸异丙酯 Isopropyl 4-Methylbenzenesulfonate 2307-69-9 9 C3D-1724 Methyl (R)-2-amino-2-phenylacetate hydrochloride Methyl (R)-2-Amino-2-Phenylacetate Hydrochloride 19883-41-1 10 C3D-1792 睾酮 Testosterone 58-22-0 11 C3D-1804 1-羟基苯并三唑 1-Hydroxybenzotriazole 2592-95-2 12 C3D-1808 苯甲醇 Benzyl alcohol 100-51-6 13 C3D-1823 2- [4-(溴甲基)苯基]丙酸 2-[4-(Bromomethyl)phenyl]propionic Acid 111128-12-2 14 C3D-1824 甲基-2-氧代环戊烷羧酸 Methyl 2-Oxocyclopentanecarboxylate 10472-24-9 15 C3D-1831 N-(2-甲基-5-硝基苯基)-4-(吡啶-3-基)嘧啶-2-胺 N-(2-Methyl-5-nitrophenyl)-4-(pyridin-3-yl)pyrimidin-2-amine 152460-09-8 16 C3D-1837 L-丙氨酰-L-谷氨酰胺 L-Alanyl-L-glutamine 39537-23-0 17 C3D-1848 1咪唑乙酸