Pro- Pro IP P DMAPP IPP PP, { GPP Synthase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Open 1-Dissertation NEK V12 4
The Pennsylvania State University The Graduate School Department of Chemical Engineering DEVELOPMENT OF BIOLOGICAL PLATFORM FOR THE AUTOTROPHIC PRODUCTION OF BIOFUELS A Dissertation in Chemical Engineering by Nymul Khan 2015 Nymul Khan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy May 2015 The dissertation of Nymul Khan was reviewed and approved* by the following: Wayne Curtis Professor, Chemical Engineering Dissertation Advisor Chair of Committee Esther Gomez Assistant Professor, Chemical Engineering Manish Kumar Assistant Professor, Chemical Engineering John Regan Professor, Environmental Engineering Phillip E. Savage Walter L. Robb Family Endowed Chair Head of the Department of Chemical Engineering *Signatures are on file in the Graduate School iii ABSTRACT The research described herein is aimed at developing an advanced biofuel platform that has the potential to surpass the natural rate of solar energy capture and CO2 fixation. The underlying concept is to use the electricity from a renewable source, such as wind or solar, to capture CO2 via a biological agent, such as a microbe, into liquid fuels that can be used for the transportation sector. In addition to being renewable, the higher rate of energy capture by photovoltaic cells than natural photosynthesis is expected to facilitate higher rate of liquid fuel production than traditional biofuel processes. The envisioned platform is part of ARPA-E’s (Advanced Research Projects Agency - Energy) Electrofuels initiative which aims at supplementing the country’s petroleum based fuel production with renewable liquid fuels that can integrate easily with the existing refining and distribution infrastructure (http://arpa- e.energy.gov/ProgramsProjects/Electrofuels.aspx). -

(12) United States Patent (10) Patent No.: US 8,535,916 B2 Del Cardayre Et Al
US008535916B2 (12) United States Patent (10) Patent No.: US 8,535,916 B2 Del Cardayre et al. (45) Date of Patent: Sep. 17, 2013 (54) MODIFIEDMICROORGANISMS AND USES (51) Int. Cl. THEREFOR CI2P 7/64 (2006.01) (52) U.S. Cl. (75) Inventors: Stephen B. Del Cardayre, Belmont, CA USPC .......................................................... 435/134 (US); Shane Brubaker, Oakland, CA (58) Field of Classification Search (US); Jay D. Keasling, Berkeley, CA USPC .......................................................... 435/134 (US) See application file for complete search history. (73) Assignee: LS9, Inc., South San Francisco, CA (US) (56) References Cited (*) Notice: Subject to any disclaimer, the term of this PUBLICATIONS patent is extended or adjusted under 35 IPER PCT/US2007/003736 (2007).* U.S.C. 154(b) by 497 days. Ohmiya et al. Journal of Bioscience and Bioengineering,95(6): 549 561 (2003).* (21) Appl. No.: 12/526,209 * cited by examiner (22) PCT Filed: Feb. 13, 2007 Primary Examiner — Tekchand Saidha (86). PCT No.: PCT/US2007/003736 (74) Attorney, Agent, or Firm — Brigitte A. Hajos S371 (c)(1), (57) ABSTRACT (2), (4) Date: Dec. 6, 2010 The invention provides a genetically modified microorgan ism that acquires the ability to consume a renewable feed (87) PCT Pub. No.: WO2008/100251 stock (such as cellulose) and produce products. This organ PCT Pub. Date: Aug. 21, 2008 ism can be used to ferment cellulose, one of the most abundant renewable resources available, and produce prod (65) Prior Publication Data uctS. US 2011 FOO97769 A1 Apr. 28, 2011 8 Claims, 1 Drawing Sheet U.S. Patent Sep. 17, 2013 US 8,535,916 B2 Anchor Peptide - Scaffoldin US 8,535,916 B2 1. -

European Patent Office U.S. Patent and Trademark Office
EUROPEAN PATENT OFFICE U.S. PATENT AND TRADEMARK OFFICE CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 The following classification changes will be effected by this Notice of Changes: Action Subclass Group(s) Symbols deleted: C12Y 101/01063 C12Y 101/01128 C12Y 101/01161 C12Y 102/0104 C12Y 102/03011 C12Y 103/01004 C12Y 103/0103 C12Y 103/01052 C12Y 103/99007 C12Y 103/9901 C12Y 103/99013 C12Y 103/99021 C12Y 105/99001 C12Y 105/99002 C12Y 113/11013 C12Y 113/12012 C12Y 114/15002 C12Y 114/99028 C12Y 204/01119 C12Y 402/01052 C12Y 402/01058 C12Y 402/0106 C12Y 402/01061 C12Y 601/01025 C12Y 603/02027 Symbols newly created: C12Y 101/01318 C12Y 101/01319 C12Y 101/0132 C12Y 101/01321 C12Y 101/01322 C12Y 101/01323 C12Y 101/01324 C12Y 101/01325 C12Y 101/01326 C12Y 101/01327 C12Y 101/01328 C12Y 101/01329 C12Y 101/0133 C12Y 101/01331 C12Y 101/01332 C12Y 101/01333 CPC Form – v.4 CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 Action Subclass Group(s) C12Y 101/01334 C12Y 101/01335 C12Y 101/01336 C12Y 101/01337 C12Y 101/01338 C12Y 101/01339 C12Y 101/0134 C12Y 101/01341 C12Y 101/01342 C12Y 101/03043 C12Y 101/03044 C12Y 101/98003 C12Y 101/99038 C12Y 102/01083 C12Y 102/01084 C12Y 102/01085 C12Y 102/01086 C12Y 103/01092 C12Y 103/01093 C12Y 103/01094 C12Y 103/01095 C12Y 103/01096 C12Y 103/01097 C12Y 103/0701 C12Y 103/08003 C12Y 103/08004 C12Y 103/08005 C12Y 103/08006 C12Y 103/08007 C12Y 103/08008 C12Y 103/08009 C12Y 103/99032 C12Y 104/01023 C12Y 104/01024 C12Y 104/03024 C12Y 105/01043 C12Y 105/01044 C12Y 105/01045 C12Y 105/03019 C12Y 105/0302 -

Elucidating the Biochemical Wizardry of Triterpene Metabolism in Botroycoccus Braunii
University of Kentucky UKnowledge Theses and Dissertations--Plant and Soil Sciences Plant and Soil Sciences 2011 ELUCIDATING THE BIOCHEMICAL WIZARDRY OF TRITERPENE METABOLISM IN BOTROYCOCCUS BRAUNII Thomas Daniel Niehaus University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Niehaus, Thomas Daniel, "ELUCIDATING THE BIOCHEMICAL WIZARDRY OF TRITERPENE METABOLISM IN BOTROYCOCCUS BRAUNII" (2011). Theses and Dissertations--Plant and Soil Sciences. 1. https://uknowledge.uky.edu/pss_etds/1 This Doctoral Dissertation is brought to you for free and open access by the Plant and Soil Sciences at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Plant and Soil Sciences by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained and attached hereto needed written permission statements(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine). I hereby grant to The University of Kentucky and its agents the non-exclusive license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. I agree that the document mentioned above may be made available immediately for worldwide access unless a preapproved embargo applies. -

Botryococcus Braunii
Fang et al. Biotechnol Biofuels (2015) 8:130 DOI 10.1186/s13068-015-0307-y RESEARCH ARTICLE Open Access Transcriptomic analysis of a moderately growing subisolate Botryococcus braunii 779 (Chlorophyta) in response to nitrogen deprivation Lei Fang1,6†, Deying Sun2,7†, Zhenyu Xu1, Jing He3, Shuyuan Qi1, Xin Chen4, Wee Chew5 and Jianhua Liu1,3* Abstract Background: The colonial microalga Botryococcus braunii has been brought to people’s attention for its conspicuous ability to accumulate a variety of lipids including hydrocarbons. B. braunii strains are classified into 3 races based on the types of hydrocarbons. A and B races are known to accumulate high level of lipids. However, their extreme slow growth rate has impeded its application for renewable biofuel production. Results: In this study, we report the transcriptomic response of a moderately growing subisolate from the culture of Botryococcus sp. CCALA-779 upon nitrogen deprivation (ND). We show that the subisolate has an average growth 1 1 rate of 0.52 g l− day− under photoautotrophic growth conditions and lipid content is enhanced to 75 % of CDW upon ND. Both rDNA sequence and hydrocarbon composition analyses indicate that the subisolate belongs to A race B. braunii. Hence, it is designated as B. braunii 779. We show that B. braunii 779 transcriptome shares homology to majority of the A race but not B race B. braunii ESTs, suggesting that transcriptomes of A race differ from that of B race. We found that many homologous ESTs between A races 779 and Bot-88 are unknown sequences, implying that A race contains many unknown genes. -

Nghiên Cứu Squalene Từ Vi Tảo Biển Dị Dƣỡng Schizochytrium Mangrovei Pq6 Định Hƣớng Làm Nguyên Liệu Cho Thực Phẩm Bảo Vệ Sức Khỏe, Mỹ Phẩm Và Dƣợc Phẩm
BỘ GIÁO DỤC VÀ ĐÀO TẠO VIỆN HÀN LÂM KHOA HỌC VÀ CÔNG NGHỆ VIỆT NAM HỌC VIỆN KHOA HỌC VÀ CÔNG NGHỆ ----------------------------- NGUYỄN CẨM HÀ NGHIÊN CỨU SQUALENE TỪ VI TẢO BIỂN DỊ DƢỠNG SCHIZOCHYTRIUM MANGROVEI PQ6 ĐỊNH HƢỚNG LÀM NGUYÊN LIỆU CHO THỰC PHẨM BẢO VỆ SỨC KHỎE, MỸ PHẨM VÀ DƢỢC PHẨM LUẬN ÁN TIẾN S SINH HỌC Hà Nội - Năm 2020 BỘ GIÁO DỤC VÀ ĐÀO TẠO VIỆN HÀN LÂM KHOA HỌC VÀ CÔNG NGHỆ VIỆT NAM HỌC VIỆN KHOA HỌC VÀ CÔNG NGHỆ ----------------------------- NGUYỄN CẨM HÀ NGHIÊN CỨU SQUALENE TỪ VI TẢO BIỂN DỊ DƢỠNG SCHIZOCHYTRIUM MANGROVEI PQ6 ĐỊNH HƢỚNG LÀM NGUYÊN LIỆU CHO THỰC PHẨM BẢO VỆ SỨC KHỎE, MỸ PHẨM VÀ DƢỢC PHẨM Chuyên ngành: Hóa sinh học Mã số: 9 42 01 16 LUẬN ÁN TIẾN S SINH HỌC NGƯỜI HƯỚNG DẪN KHOA HỌC: GS. TS Đặng Diễm Hồng Hà Nội – Năm 2020 i LỜI CẢM ƠN Lời đầu tiên, tôi xin được bày tỏ lòng kính trọng và biết ơn sâu sắc tới GS. TS. Đặng Diễm Hồng, nguyên Trưởng phòng Công nghệ tảo, Viện Công nghệ sinh học, Viện Hàn lâm Khoa học và Công nghệ Việt Nam - người thầy đã định hướng, truyền dạy những kiến thức khoa học và giúp đỡ tôi vượt qua những trở ngại và khó khăn trong suốt thời gian thực hiện luận án. Tôi trân trọng cảm ơn Ban Lãnh đạo Viện Công nghệ sinh học, phòng thí nghiệm trọng điểm Công nghệ gen, bộ phận đào tạo Viện Công nghệ sinh học, Ban Lãnh đạo và bộ phận đào tạo của Học viện Khoa học và Công nghệ, Viện Hàn lâm Khoa học và Công nghệ Việt Nam đã tạo điều kiện thuận lợi về cơ sở vật chất và giúp tôi hoàn thành mọi thủ tục cần thiết trong quá trình làm nghiên cứu. -

Ppor-L's Pports IPP DMAPP
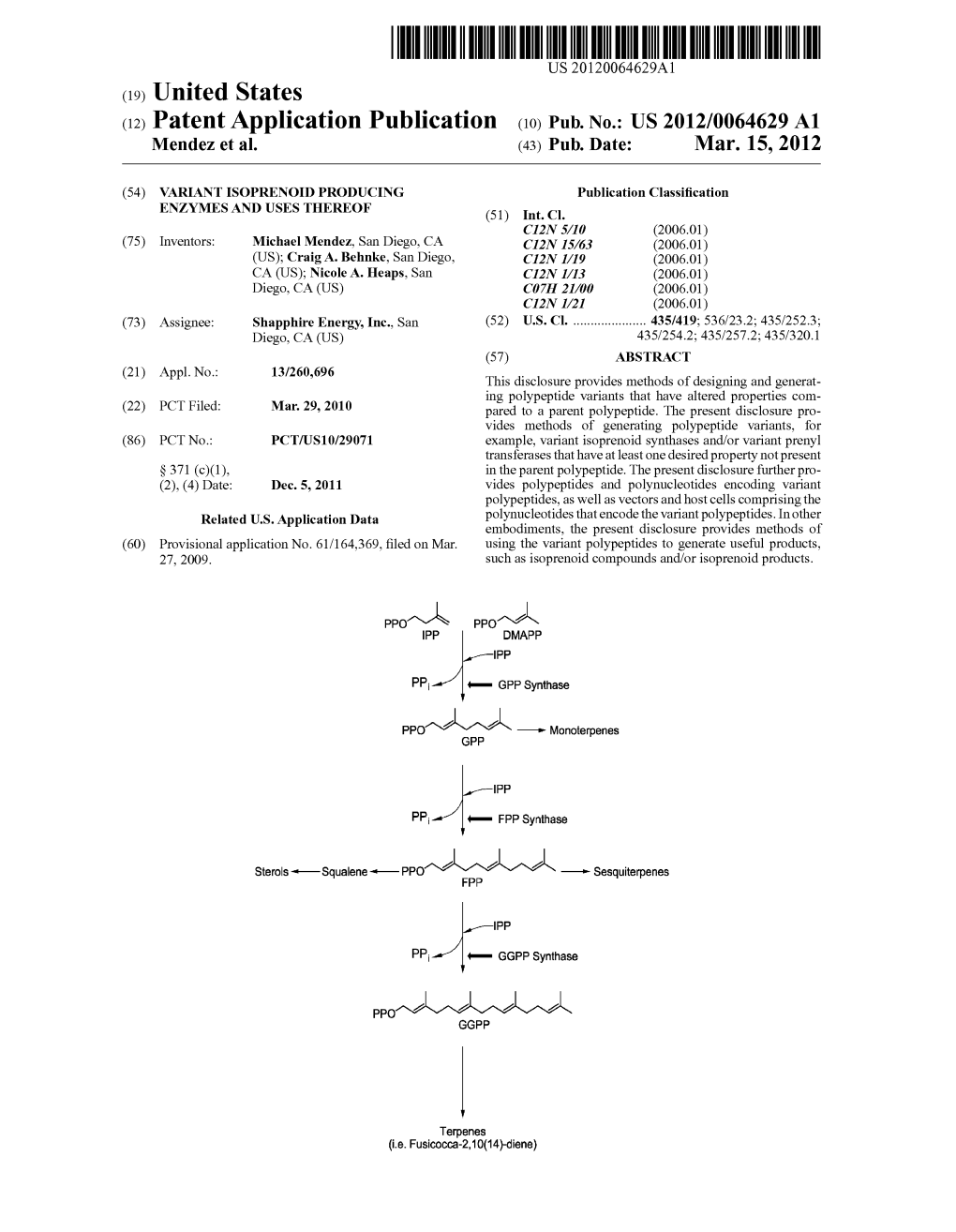
US 20120058535A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0058535 A1 Heaps et al. (43) Pub. Date: Mar. 8, 2012 (54) BIOFUEL PRODUCTION IN PROKARYOTES Publication Classification AND EUKARYOTES (51) Int. Cl. (75) Inventors: Nicole A. Heaps, San Diego, CA CI2N 9/00 (2006.01) (US); Craig A. Behnke, San Diego, CI2N I/2 (2006.01) CA (US); David Molina, San CI2N 15/63 (2006.01) Diego, CA (US) CI2N I/3 (2006.01) CI2N 5/10 (2006.01) (73) Assignee: SAPPHIRE ENERGY, INC., SAN C7H 2L/04 (2006.01) DIEGO, CA (US) CI2N I/19 (2006.01) (21) Appl. No.: 13/255,888 (52) U.S. Cl. ..................... 435/183: 536/23.1: 435/252.3: 435/254.2:435/257.2:435/419:435/320.1 (22) PCT Filed: Mar. 5, 2010 (86). PCT No.: PCT/US 10/26445 (57) ABSTRACT S371 (c)(1), Terpene synthases are enzymes that directly convert IPP & DMAPP to terpenes, such as fusicoccadiene. Described (2), (4) Date: Nov. 9, 2011 herein are methods and compositions for the production of (30) Foreign Application Priority Data terpenes and terpenoids for use as fuel molecules or other useful components. Genetically engineered enzymes capable Mar. 11, 2009 (US) .................................... 61159366 of producing terpenes and terpenoids are also described. ppor-l's pports IPP DMAPP IPP ppor--~s - Monoterpenes GPP PP Sterols --Squalene-PPO 2 2 1N -- Sesquiterpenes FPP PP PPO1 na 2 2 GGPP Terpenes (i.e. Fusicocca-2,10(14)-diene) Patent Application Publication Mar. 8, 2012 Sheet 1 of 32 US 2012/0058535 A1 ppor-l's pports IPP DMAPP PP ppor--~s Monoterpenes GPP IPP Sterols -HSqualene-HPPO 2 FSG 2 Sesquiterpenes PP PPO 12 12 1. -

In Silico Modeling and Characterization of Squalene
Environ & m m en u ta le l o B r t i o e t Elumalai et al., J Pet Environ Biotechnol 2018, 9:3 P e f c h o Journal of Petroleum & n l o a D0I: 10.4172/2157-7463.1000371 l n o r g u y o J Environmental Biotechnology ISSN: 2157-7463 Research Article Open Access In Silico Modeling and Characterization of Squalene Synthase and Botryococcene Synthase Enzymes from a Green Photosynthetic Microalga Botryococcus braunii Elumalai S1*, Sangeetha T1 and Rajesh Kanna G2 1Department of Biotechnology, University of Madras, Guindy Campus, Chennai, India 2Department of Plant Biology and Plant Biotechnology, Presidency College (Autonomous), Chennai, India Abstract The green photosynthetic microalgae are considered as a major source of lipids from lacustrine and marine environments. Among them, Botryococcus braunii plays a key role due to its high efficiency production of huge amount of unsaturated hydrocarbons up to 75% of its dry weight. Evidently, more number of new compounds has been reported in sediments as deposits in both the marine and lacustrine environments. These depositions are reported as sediments from the algal lipids during the course of evolution. Botryococcene is one among the long chain hydrocarbon reported in higher amount extracellular as depositions from this microalga Botryococcus braunii race B. However, mass cultivation of this microalga for botryococcene as sustainable and renewable biofuel is a challenging target due to its doubling time and slow growth. Therefore, genetic engineering may play a key role to solve this issue. In addition to squalene synthase, squalene synthase-like genes have been reported from the race B of B. -
Metabolic Engineering and Systems Biology for Increasing Biofuel Production in Microalgae
METABOLIC ENGINEERING AND SYSTEMS BIOLOGY FOR INCREASING BIOFUEL PRODUCTION IN MICROALGAE by Robert Edward Jinkerson Copyright by Robert E. Jinkerson 2013 Some Rights Reserved This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License http://creativecommons.org/licenses/by-nc-sa/4.0/ A thesis submitted to the Faculty and the Board of Trustees of the Colorado School of Mines in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Applied Chemistry). Golden, Colorado Date ___________ Signed: _______________________ Robert Jinkerson Signed: _______________________ Dr. Matthew C. Posewitz Thesis Advisor Golden, Colorado Date ___________ Signed: _______________________ Dr. David T. Wu Professer and Head Department of Chemistry and Geochemistry ii ABSTRACT The saccharification of starch coupled with fermentation to ethanol and the transesterification of vegetable oils to produce biodiesel are mature technologies that currently provide the majority of biofuels in the United States. However, traditional food-based starch and oil feedstocks, such as corn and soybean, cannot meet our current fuel demands and their use remains controversial. Microalgae have been of recent interest for use as a biofuel feedstock because they can produce large quantities of carbohydrates and lipids, contain little recalcitrant biomass, and do not impact the food supply. In the green alga Chlamydomonas reinhardtii, the primary carbohydrate produced is starch and the primary storage lipid produced is triacylglyceride (TAG), but high yields of these bioenergy carriers are usually only found under conditions of nutrient stress (N, S, P). As a strategy for increasing TAG yields, starchless mutants that divert carbon away from starch biosynthesis and into TAG production were investigated. -

Conference Committees
Conference Committees Executive Committee Prof. Dr. Ghulamqadir Kazi Prof. Dr. Mehmet Karatas Prof. Dr. Umar Ali Khan Dr. Muhammad Aasim Dr. Zahid Iqbal Organizing Committee Prof. Dr. Umar Ali Khan Prof. Dr. lshtiaq Ahmed Dr. Zahid Iqbal Prof. Dr. Ghulam Mustafa Lodhi Mr. Mustafa Minhas Mr. Aafaq Ahmed Technical Committee Prof. Dr. Mehmet Karatas - Turkey Prof. Dr. Raheel Qamar - Pakistan Prof. Dr. lshtiaq Ahmed - Pakistan Prof. Dr. Aamir Ali Khan - Pakistan Prof. Dr. ljaz Javed Hasan - Pakistan Prof. Dr. Zafar Iqbal- Pakistan Dr. Muhammad Aasim - Turkey Dr. Munir Ahmad-Pakistan Dr. Muhammad Asif Aziz - Pakistan Dr. Awais Rasheed-China Dr. Riaz Hussain - Pakistan Dr. Muhammad Aamer Mehmood, Pakistan Dr. Naeem Asghar - Pakistan Dr. Faqir Muhammad- USA Dr. Khalid F. Salamat - UK Dr. Mazhar ul Haq - Pakistan Dr. Mansur Abdullah - Pakistan Dr. Muhammad Kashif Saleemi - Pakistan Dr. Zahid Iqbal-Pakistan Designing & Publication Mr. Muhammad Imran Mr. Muhammad Laiq Khan Asian J Agri Biol, Special Issue-2015 ANTIFUNGAL POTENTIAL OF PLANT EXTRACTS FOR THE MANAGEMENT OF ANTHRACNOSE OF CHILI CAUSED BY COLLETOTRICHUM CAPSICI Muhammad Nasir Subhani1*, Muhammad Bilal Chattha1 and Waseem Abbas2 1Institute of Agricultural Sciences, University of the Punjab, Lahore, Pakistan 2Arid Zone Research Institute, Bhakar, Pakistan ABSTRACT Chili anthracnose damages chili fruits extensively at pre- and postharvest stages causing anthracnose lesions. Even very small lesion of anthracnose on fruits of chili reduces the market value of chili crop. Fungitoxic effects of methanolic plant extracts were tested in vitrothrough poisoned food technique. There was a significant decrease in mycellial growth of the fungus with an increase in methanolic plant extracts concentration in all the tested methanolic plant extracts over the control. -

Approaches in the Photosynthetic Production of Sustainable Fuels By
Approaches in the Photosynthetic Production of Sustainable Fuels by Cyanobacteria using Tools of Synthetic Biology Indrajeet .1, Akhil Rautela1, and Sanjay Kumar1 1IIT BHU July 24, 2021 Abstract Cyanobacteria, photosynthetic prokaryotic microorganisms having a simple genetic composition are the prospective photoau- totrophic cell factories for the production of a wide range of biofuel molecules. Simple genetic composition of cyanobacteria allows effortless genetic manipulation which leads to increased research endeavour from the synthetic biology approach. An improved development of synthetic biology tools, genetic modification methods and advancement in transformation techniques to construct a strain which can contain multiple target genes in single operon will vastly expand the functions that can be used for engineering photosynthetic cyanobacteria for the generation of biofuels. In this review, recent advancements and approaches in synthetic biology tools and biofuel production by metabolically engineered cyanobacteria have been discussed. Various fuel molecules like isoprene, limonene, a-farnesene, squalene, alkanes, butanol and fatty acids which can be a substitute of petroleum and fossil fuels in future have been elaborated. Introduction Industrialization and the human population explosion have created a huge energy crisis worldwide due to their dependence on the non-conventional fossil fuels like petroleum, coal and natural gases to fulfil their daily energy requirements (Chandrasekhar et al., 2015; Kumar et al., 2017; Pandey et al., 2020). Extensive use of fossil fuels also generates a huge amount of harmful greenhouse gases which get accumulated in the environment and adversely affect the life on the planet. Harvesting solar energy via photosynthesis is one of nature's noteworthy achievements that could also be a solution for the future world-wide energy need. -

Lipidomics for Geochemistry
This article was originally published in Treatise on Geochemistry, Second Edition published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non- commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who you know, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial Pearson A. (2014) Lipidomics for Geochemistry. In: Holland H.D. and Turekian K.K. (eds.) Treatise on Geochemistry, Second Edition, vol. 12, pp. 291-336. Oxford: Elsevier. © 2014 Elsevier Ltd. All rights reserved. Author's personal copy 12.11 Lipidomics for Geochemistry A Pearson, Harvard University, Cambridge, MA, USA ã 2014 Elsevier Ltd. All rights reserved. 12.11.1 Introduction 291 12.11.2 Lipid Biosynthetic Pathways 292 12.11.2.1 Acetogenic Lipids 292 12.11.2.2 Isoprenoids – MVA Pathway 295 12.11.2.3 Isoprenoids – MEP Pathway 296 12.11.2.4 Ether and Ester Linkages to Glycerol – Bacteria and Eukaryotes 297 12.11.2.5 Diethers and Tetraethers of Archaea 300 12.11.2.6 Glycerol Membrane Lipids: Final Comments 302 12.11.2.7