Algae for Biofuels and Energy Developments in Applied Phycology 5
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Template for Two-Page Abstracts in Word 97 (PC)
GEOLOGIC MAPPING OF THE LUNAR SOUTH POLE QUADRANGLE (LQ-30). S.C. Mest1,2, D.C. Ber- man1, and N.E. Petro2, 1Planetary Science Institute, 1700 E. Ft. Lowell, Suite 106, Tucson, AZ 85719-2395 ([email protected]); 2Planetary Geodynamics Laboratory, Code 698, NASA GSFC, Greenbelt, MD 20771. Introduction: In this study we use recent image, the surface [7]. Impact craters display morphologies spectral and topographic data to map the geology of the ranging from simple to complex [7-9,24] and most lunar South Pole quadrangle (LQ-30) at 1:2.5M scale contain floor deposits distinct from surrounding mate- [1-7]. The overall objective of this research is to con- rials. Most of these deposits likely consist of impact strain the geologic evolution of LQ-30 (60°-90°S, 0°- melt; however, some deposits, especially on the floors ±180°) with specific emphasis on evaluation of a) the of the larger craters and basins (e.g., Antoniadi), ex- regional effects of impact basin formation, and b) the hibit low albedo and smooth surfaces and may contain spatial distribution of ejecta, in particular resulting mare. Higher albedo deposits tend to contain a higher from formation of the South Pole-Aitken (SPA) basin density of superposed impact craters. and other large basins. Key scientific objectives in- Antoniadi Crater. Antoniadi crater (D=150 km; clude: 1) Determining the geologic history of LQ-30 69.5°S, 172°W) is unique for several reasons. First, and examining the spatial and temporal variability of Antoniadi is the only lunar crater that contains both a geologic processes within the map area. -

Methanothermus Fervidus Type Strain (V24S)
UC Davis UC Davis Previously Published Works Title Complete genome sequence of Methanothermus fervidus type strain (V24S). Permalink https://escholarship.org/uc/item/9367m39j Journal Standards in genomic sciences, 3(3) ISSN 1944-3277 Authors Anderson, Iain Djao, Olivier Duplex Ngatchou Misra, Monica et al. Publication Date 2010-11-20 DOI 10.4056/sigs.1283367 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Standards in Genomic Sciences (2010) 3:315-324 DOI:10.4056/sigs.1283367 Complete genome sequence of Methanothermus fervidus type strain (V24ST) Iain Anderson1, Olivier Duplex Ngatchou Djao2, Monica Misra1,3, Olga Chertkov1,3, Matt Nolan1, Susan Lucas1, Alla Lapidus1, Tijana Glavina Del Rio1, Hope Tice1, Jan-Fang Cheng1, Roxanne Tapia1,3, Cliff Han1,3, Lynne Goodwin1,3, Sam Pitluck1, Konstantinos Liolios1, Natalia Ivanova1, Konstantinos Mavromatis1, Natalia Mikhailova1, Amrita Pati1, Evelyne Brambilla4, Amy Chen5, Krishna Palaniappan5, Miriam Land1,6, Loren Hauser1,6, Yun-Juan Chang1,6, Cynthia D. Jeffries1,6, Johannes Sikorski4, Stefan Spring4, Manfred Rohde2, Konrad Eichinger7, Harald Huber7, Reinhard Wirth7, Markus Göker4, John C. Detter1, Tanja Woyke1, James Bristow1, Jonathan A. Eisen1,8, Victor Markowitz5, Philip Hugenholtz1, Hans-Peter Klenk4, and Nikos C. Kyrpides1* 1 DOE Joint Genome Institute, Walnut Creek, California, USA 2 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 5 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 6 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 7 University of Regensburg, Archaeenzentrum, Regensburg, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Nikos C. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Perspectives in Phycology Vol
Perspectives in Phycology Vol. 3 (2016), Issue 3, p. 141–154 Article Published online June 2016 Diversity and ecology of green microalgae in marine systems: an overview based on 18S rRNA gene sequences Margot Tragin1, Adriana Lopes dos Santos1, Richard Christen2,3 and Daniel Vaulot1* 1 Sorbonne Universités, UPMC Univ Paris 06, CNRS, UMR 7144, Station Biologique, Place Georges Teissier, 29680 Roscoff, France 2 CNRS, UMR 7138, Systématique Adaptation Evolution, Parc Valrose, BP71. F06108 Nice cedex 02, France 3 Université de Nice-Sophia Antipolis, UMR 7138, Systématique Adaptation Evolution, Parc Valrose, BP71. F06108 Nice cedex 02, France * Corresponding author: [email protected] With 5 figures in the text and an electronic supplement Abstract: Green algae (Chlorophyta) are an important group of microalgae whose diversity and ecological importance in marine systems has been little studied. In this review, we first present an overview of Chlorophyta taxonomy and detail the most important groups from the marine environment. Then, using public 18S rRNA Chlorophyta sequences from culture and natural samples retrieved from the annotated Protist Ribosomal Reference (PR²) database, we illustrate the distribution of different green algal lineages in the oceans. The largest group of sequences belongs to the class Mamiellophyceae and in particular to the three genera Micromonas, Bathycoccus and Ostreococcus. These sequences originate mostly from coastal regions. Other groups with a large number of sequences include the Trebouxiophyceae, Chlorophyceae, Chlorodendrophyceae and Pyramimonadales. Some groups, such as the undescribed prasinophytes clades VII and IX, are mostly composed of environmental sequences. The 18S rRNA sequence database we assembled and validated should be useful for the analysis of metabarcode datasets acquired using next generation sequencing. -

Open 1-Dissertation NEK V12 4
The Pennsylvania State University The Graduate School Department of Chemical Engineering DEVELOPMENT OF BIOLOGICAL PLATFORM FOR THE AUTOTROPHIC PRODUCTION OF BIOFUELS A Dissertation in Chemical Engineering by Nymul Khan 2015 Nymul Khan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy May 2015 The dissertation of Nymul Khan was reviewed and approved* by the following: Wayne Curtis Professor, Chemical Engineering Dissertation Advisor Chair of Committee Esther Gomez Assistant Professor, Chemical Engineering Manish Kumar Assistant Professor, Chemical Engineering John Regan Professor, Environmental Engineering Phillip E. Savage Walter L. Robb Family Endowed Chair Head of the Department of Chemical Engineering *Signatures are on file in the Graduate School iii ABSTRACT The research described herein is aimed at developing an advanced biofuel platform that has the potential to surpass the natural rate of solar energy capture and CO2 fixation. The underlying concept is to use the electricity from a renewable source, such as wind or solar, to capture CO2 via a biological agent, such as a microbe, into liquid fuels that can be used for the transportation sector. In addition to being renewable, the higher rate of energy capture by photovoltaic cells than natural photosynthesis is expected to facilitate higher rate of liquid fuel production than traditional biofuel processes. The envisioned platform is part of ARPA-E’s (Advanced Research Projects Agency - Energy) Electrofuels initiative which aims at supplementing the country’s petroleum based fuel production with renewable liquid fuels that can integrate easily with the existing refining and distribution infrastructure (http://arpa- e.energy.gov/ProgramsProjects/Electrofuels.aspx). -

University of Oklahoma
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By JOSHUA THOMAS COOPER Norman, Oklahoma 2017 MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Boris Wawrik, Chair ______________________________ Dr. J. Phil Gibson ______________________________ Dr. Anne K. Dunn ______________________________ Dr. John Paul Masly ______________________________ Dr. K. David Hambright ii © Copyright by JOSHUA THOMAS COOPER 2017 All Rights Reserved. iii Acknowledgments I would like to thank my two advisors Dr. Boris Wawrik and Dr. J. Phil Gibson for helping me become a better scientist and better educator. I would also like to thank my committee members Dr. Anne K. Dunn, Dr. K. David Hambright, and Dr. J.P. Masly for providing valuable inputs that lead me to carefully consider my research questions. I would also like to thank Dr. J.P. Masly for the opportunity to coauthor a book chapter on the speciation of diatoms. It is still such a privilege that you believed in me and my crazy diatom ideas to form a concise chapter in addition to learn your style of writing has been a benefit to my professional development. I’m also thankful for my first undergraduate research mentor, Dr. Miriam Steinitz-Kannan, now retired from Northern Kentucky University, who was the first to show the amazing wonders of pond scum. Who knew that studying diatoms and algae as an undergraduate would lead me all the way to a Ph.D. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Histone Variants in Archaea and the Evolution of Combinatorial Chromatin Complexity
Histone variants in archaea and the evolution of combinatorial chromatin complexity Kathryn M. Stevensa,b, Jacob B. Swadlinga,b, Antoine Hochera,b, Corinna Bangc,d, Simonetta Gribaldoe, Ruth A. Schmitzc, and Tobias Warneckea,b,1 aMolecular Systems Group, Quantitative Biology Section, Medical Research Council London Institute of Medical Sciences, London W12 0NN, United Kingdom; bInstitute of Clinical Sciences, Faculty of Medicine, Imperial College London, London W12 0NN, United Kingdom; cInstitute for General Microbiology, University of Kiel, 24118 Kiel, Germany; dInstitute of Clinical Molecular Biology, University of Kiel, 24105 Kiel, Germany; and eDepartment of Microbiology, Unit “Evolutionary Biology of the Microbial Cell,” Institut Pasteur, 75015 Paris, France Edited by W. Ford Doolittle, Dalhousie University, Halifax, NS, Canada, and approved October 28, 2020 (received for review April 14, 2020) Nucleosomes in eukaryotes act as platforms for the dynamic inte- additional histone dimers can be taggedontothistetramertoyield gration of epigenetic information. Posttranslational modifications oligomers of increasing length that wrap correspondingly more DNA are reversibly added or removed and core histones exchanged for (3, 6–9). Almost all archaeal histones lack tails and PTMs have yet to paralogous variants, in concert with changing demands on tran- be reported. Many archaea do, however, encode multiple histone scription and genome accessibility. Histones are also common in paralogs (8, 10) that can flexibly homo- and heterodimerize in -
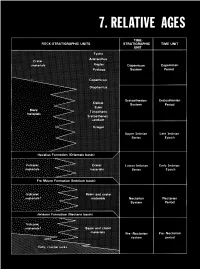
Relative Ages
CONTENTS Page Introduction ...................................................... 123 Stratigraphic nomenclature ........................................ 123 Superpositions ................................................... 125 Mare-crater relations .......................................... 125 Crater-crater relations .......................................... 127 Basin-crater relations .......................................... 127 Mapping conventions .......................................... 127 Crater dating .................................................... 129 General principles ............................................. 129 Size-frequency relations ........................................ 129 Morphology of large craters .................................... 129 Morphology of small craters, by Newell J. Fask .................. 131 D, method .................................................... 133 Summary ........................................................ 133 table 7.1). The first three of these sequences, which are older than INTRODUCTION the visible mare materials, are also dominated internally by the The goals of both terrestrial and lunar stratigraphy are to inte- deposits of basins. The fourth (youngest) sequence consists of mare grate geologic units into a stratigraphic column applicable over the and crater materials. This chapter explains the general methods of whole planet and to calibrate this column with absolute ages. The stratigraphic analysis that are employed in the next six chapters first step in reconstructing -

An Unrecognized Ancient Lineage of Green Plants Persists in Deep Marine Waters1
J. Phycol. 46, 1288–1295 (2010) Ó 2010 Phycological Society of America DOI: 10.1111/j.1529-8817.2010.00900.x AN UNRECOGNIZED ANCIENT LINEAGE OF GREEN PLANTS PERSISTS IN DEEP MARINE WATERS1 Frederick W. Zechman2,3 Department of Biology, California State University Fresno, 2555 East San Ramon Ave, Fresno, California 93740, USA Heroen Verbruggen,3 Frederik Leliaert Phycology Research Group, Ghent University, Krijgslaan 281 S8, 9000 Ghent, Belgium Matt Ashworth University Station MS A6700, 311 Biological Laboratories, University of Texas at Austin, Austin, Texas 78712, USA Mark A. Buchheim Department of Biological Science, University of Tulsa, Tulsa, Oklahoma 74104, USA Marvin W. Fawley School of Mathematical and Natural Sciences, University of Arkansas at Monticello, Monticello, Arkansas 71656, USA Department of Biological Sciences, North Dakota State University, Fargo, North Dakota 58105, USA Heather Spalding Botany Department, University of Hawaii at Manoa, Honolulu, Hawaii 96822, USA Curt M. Pueschel Department of Biological Sciences, State University of New York at Binghamton, Binghamton, New York 13901, USA Julie A. Buchheim, Bindhu Verghese Department of Biological Science, University of Tulsa, Tulsa, Oklahoma 74104, USA and M. Dennis Hanisak Harbor Branch Oceanographic Institution, Fort Pierce, Florida 34946, USA We provide molecular phylogenetic evidence that Key index words: Chlorophyta; green algae; molec- the obscure genera Palmophyllum Ku¨tz. and Verdigel- ular phylogenetics; Palmophyllaceae fam. nov.; las D. L. Ballant. et J. N. Norris form a distinct and Palmophyllales ord. nov.; Palmophyllum; Prasino- early diverging lineage of green algae. These pal- phyceae; Verdigellas; Viridiplantae melloid seaweeds generally persist in deep waters, Abbreviations: AU, approximately unbiased; BI, where grazing pressure and competition for space Bayesian inference; ML, maximum likelihood; are reduced. -

Prokaryotic Biodiversity of Lonar Meteorite Crater Soda Lake Sediment and Community Dynamics During Microenvironmental Ph Homeostasis by Metagenomics
Prokaryotic Biodiversity of Lonar Meteorite Crater Soda Lake Sediment and Community Dynamics During Microenvironmental pH Homeostasis by Metagenomics Dissertation for the award of the degree "Doctor of Philosophy" Ph.D. Division of Mathematics and Natural Sciences of the Georg-August-Universität Göttingen within the doctoral program in Biology of the Georg-August University School of Science (GAUSS) Submitted by Soumya Biswas from Ranchi (India) Göttingen, 2016 Thesis Committee Prof. Dr. Rolf Daniel Department of Genomic and Applied Microbiology, Institute of Microbiology and Genetics, Faculty of Biology and Psychology, Georg-August-Universität Göttingen, Germany PD Dr. Michael Hoppert Department of General Microbiology, Institute of Microbiology and Genetics, Faculty of Biology and Psychology, Georg-August-Universität Göttingen, Germany Members of the Examination Board Reviewer: Prof. Dr. Rolf Daniel, Department of Genomic and Applied Microbiology, Institute of Microbiology and Genetics, Faculty of Biology and Psychology, Georg-August-Universität Göttingen, Germany Second Reviewer: PD Dr. Michael Hoppert, Department of General Microbiology, Institute of Microbiology and Genetics, Faculty of Biology and Psychology, Georg-August-Universität Göttingen, Germany Further members of the Examination Board: Prof. Dr. Burkhard Morgenstern, Department of Bioinformatics, Institute of Microbiology and Genetics, Faculty of Biology and Psychology, Georg-August-Universität Göttingen, Germany PD Dr. Fabian Commichau, Department of General Microbiology, -

Representatives of a Novel Archaeal Phylum Or a Fast-Evolving
Open Access Research2005BrochieretVolume al. 6, Issue 5, Article R42 Nanoarchaea: representatives of a novel archaeal phylum or a comment fast-evolving euryarchaeal lineage related to Thermococcales? Celine Brochier*, Simonetta Gribaldo†, Yvan Zivanovic‡, Fabrice Confalonieri‡ and Patrick Forterre†‡ Addresses: *EA EGEE (Evolution, Génomique, Environnement) Université Aix-Marseille I, Centre Saint-Charles, 3 Place Victor Hugo, 13331 Marseille, Cedex 3, France. †Unite Biologie Moléculaire du Gène chez les Extremophiles, Institut Pasteur, 25 rue du Dr Roux, 75724 Paris Cedex ‡ 15, France. Institut de Génétique et Microbiologie, UMR CNRS 8621, Université Paris-Sud, 91405 Orsay, France. reviews Correspondence: Celine Brochier. E-mail: [email protected]. Simonetta Gribaldo. E-mail: [email protected] Published: 14 April 2005 Received: 3 December 2004 Revised: 10 February 2005 Genome Biology 2005, 6:R42 (doi:10.1186/gb-2005-6-5-r42) Accepted: 9 March 2005 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2005/6/5/R42 reports © 2005 Brochier et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Placement<p>Anteins from analysis 25of Nanoarcheumarchaeal of the positiongenomes equitans of suggests Nanoarcheum in the that archaeal N. equitans phylogeny inis likethe lyarchaeal to be the phylogeny representative using aof large a fast-evolving dataset of concatenatedeuryarchaeal ribosomalineage.</p>l pro- deposited research Abstract Background: Cultivable archaeal species are assigned to two phyla - the Crenarchaeota and the Euryarchaeota - by a number of important genetic differences, and this ancient split is strongly supported by phylogenetic analysis.