Table S1. Complete Gene Expression Data from Human Diabetes RT² Profiler™ PCR Array Receptors, Transporters & Channels* A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Title of the Dissertation
UNIVERSITY OF CALIFORNIA SAN DIEGO Novel network-based integrated analyses of multi-omics data reveal new insights into CD8+ T cell differentiation and mouse embryogenesis A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioinformatics and Systems Biology by Kai Zhang Committee in charge: Professor Wei Wang, Chair Professor Pavel Arkadjevich Pevzner, Co-Chair Professor Vineet Bafna Professor Cornelis Murre Professor Bing Ren 2018 Copyright Kai Zhang, 2018 All rights reserved. The dissertation of Kai Zhang is approved, and it is accept- able in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California San Diego 2018 iii EPIGRAPH The only true wisdom is in knowing you know nothing. —Socrates iv TABLE OF CONTENTS Signature Page ....................................... iii Epigraph ........................................... iv Table of Contents ...................................... v List of Figures ........................................ viii List of Tables ........................................ ix Acknowledgements ..................................... x Vita ............................................. xi Abstract of the Dissertation ................................. xii Chapter 1 General introduction ............................ 1 1.1 The applications of graph theory in bioinformatics ......... 1 1.2 Leveraging graphs to conduct integrated analyses .......... 4 1.3 References .............................. 6 Chapter 2 Systematic -

Regulation of Adult Neurogenesis in Mammalian Brain
International Journal of Molecular Sciences Review Regulation of Adult Neurogenesis in Mammalian Brain 1,2, 3, 3,4 Maria Victoria Niklison-Chirou y, Massimiliano Agostini y, Ivano Amelio and Gerry Melino 3,* 1 Centre for Therapeutic Innovation (CTI-Bath), Department of Pharmacy & Pharmacology, University of Bath, Bath BA2 7AY, UK; [email protected] 2 Blizard Institute of Cell and Molecular Science, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London E1 2AT, UK 3 Department of Experimental Medicine, TOR, University of Rome “Tor Vergata”, 00133 Rome, Italy; [email protected] (M.A.); [email protected] (I.A.) 4 School of Life Sciences, University of Nottingham, Nottingham NG7 2HU, UK * Correspondence: [email protected] These authors contributed equally to this work. y Received: 18 May 2020; Accepted: 7 July 2020; Published: 9 July 2020 Abstract: Adult neurogenesis is a multistage process by which neurons are generated and integrated into existing neuronal circuits. In the adult brain, neurogenesis is mainly localized in two specialized niches, the subgranular zone (SGZ) of the dentate gyrus and the subventricular zone (SVZ) adjacent to the lateral ventricles. Neurogenesis plays a fundamental role in postnatal brain, where it is required for neuronal plasticity. Moreover, perturbation of adult neurogenesis contributes to several human diseases, including cognitive impairment and neurodegenerative diseases. The interplay between extrinsic and intrinsic factors is fundamental in regulating neurogenesis. Over the past decades, several studies on intrinsic pathways, including transcription factors, have highlighted their fundamental role in regulating every stage of neurogenesis. However, it is likely that transcriptional regulation is part of a more sophisticated regulatory network, which includes epigenetic modifications, non-coding RNAs and metabolic pathways. -

A Simple Differentiation Protocol for Generation of Induced
1 Article, Special Issue "Induced Pluripotent Stem Cells in Neurodegenerative Diseases: Application for Therapy and 2 Disease Modeling" 3 A simple differentiation protocol for generation of 4 induced pluripotent stem cell-derived basal forebrain 5 cholinergic neurons for Alzheimer’s disease and 6 frontotemporal dementia disease modeling 7 Supplemental information 8 Method 1. Reprogramming and characterisation of MBE2960 healthy control iPSC line 9 The iPSCs were generated using skin fibroblasts obtained from subjects over the age of 18 years 10 by episomal method as described [40]. Briefly, reprogramming was performed on passage 8-10 11 fibroblasts by nucleofection (Lonza Amaxa Nucleofector) with episomal vectors expressing 12 OCT4, SOX2, KLF4, L-MYC, LIN28 and shRNA against p53 [41] in feeder- and serum- free 13 conditions using TeSR-E7 medium (Stemcell Technologies). Subsequently, reprogrammed 14 colonies were manually dissected to establish clonal cell lines [42]. Three clones were assessed 15 for pluripotency markers via immunocytochemistry (Figure S1A). The iPSC line was expanded 16 and characterised. Embryoid bodies were obtained as described [43] and using tri-lineage 17 differentiation kit (Stemcell Technologies). Germ layer differentiation was assessed by 18 immunochemistry (Figure S1B). Copy number variation (CNV) analysis of original fibroblasts 19 and iPSCs from MBE2960 (Figure S1C) was performed using Illumina HumanCore Beadchip 20 arrays as we described [40]. CNV analyses were performed using PennCNV and QuantiSNP with 21 default parameter settings [44,45]. Chromosomal aberrations were deemed to involve at least 10 22 contiguous single nucleotide polymorphisms (SNPs) or a genomic region spanning at least 1MB 23 [44,45]. The B allele frequency (BAF) and the log R ratio (LRR) were extracted from 24 GenomeStudio (Illumina) for representation (Figure S1D). -

Diagnosis of FOXG1 Syndrome Caused by Recurrent Balanced Chromosomal Rearrangements: Case Study and Literature Review Connor P
Craig et al. Mol Cytogenet (2020) 13:40 https://doi.org/10.1186/s13039-020-00506-1 CASE REPORT Open Access Diagnosis of FOXG1 syndrome caused by recurrent balanced chromosomal rearrangements: case study and literature review Connor P. Craig1,2, Emily Calamaro3, Chin‑To Fong3, Anwar M. Iqbal1, Alexander R. Paciorkowski3,4,5,6 and Bin Zhang1,3* Abstract Background: The FOXG1 gene plays a vital role in mammalian brain diferentiation and development. Intra‑ and intergenic mutations resulting in loss of function or altered expression of the FOXG1 gene cause FOXG1 syndrome. The hallmarks of this syndrome are severe developmental delay with absent verbal language, post‑natal growth restric‑ tion, post‑natal microcephaly, and a recognizable movement disorder characterized by chorea and dystonia. Case presentation: Here we describe a case of a 7‑year‑old male patient found to have a de novo balanced translo‑ cation between chromosome 3 at band 3q14.1 and chromosome 14 at band 14q12 via G‑banding chromosome and Fluorescence In Situ Hybridization (FISH) analyses. This rearrangement disrupts the proximity of FOXG1 to a previously described smallest region of deletion overlap (SRO), likely resulting in haploinsufciency. Conclusions: This case adds to the growing body of literature implicating chromosomal structural variants in the manifestation of this disorder and highlights the vital role of cis‑acting regulatory elements in the normal expression of this gene. Finally, we propose a protocol for refex FISH analysis to improve diagnostic efciency for patients with suspected FOXG1 syndrome. Keywords: FOXG1, Haploinsufciency, Postnatal microcephaly, FISH, Enhancer, Chromosomal rearrangement, Diagnosis Introduction brain development, with high levels of expression in the Te Forkhead Box G1 (FOXG1) gene [OMIM: 164874], developing fetal telencephalon [1–4]. -
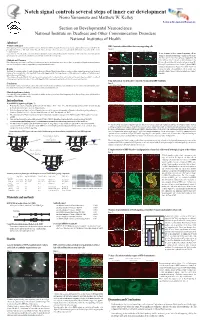
Notch Signal Controls Several Steps of Inner Ear Development Norio Yamamoto and Matthew W
NIDCD Notch signal controls several steps of inner ear development Norio Yamamoto and Matthew W. Kelley Section on Developmental Neuroscience Section on Developmental Neuroscience National Institute on Deafness and Other Communication Disorders National Institutes of Health Abstract Problem addressed RBP-J mutant cochleae did not have any supporting cells Notch signaling has been reported to contribute to inner ear development, however, its specific functions remain unclear, partly because of discrepancies between the phenotypes of mutant mice with single deletion of specific Notch related genes. These discrepancies are probably due to functional compensation by other Notch Figure 4 receptors or ligands. Foxg1 Cre;RBP-J floxed/+ Foxg1 Cre;RBP-J floxed/floxed A B C D To determine the effects of complete elimination of Notch signaling, we used a conditional knockout of the Rbpsuh gene. RBP-J protein is a critical transcriptional p27 Phalloidin p27 Phalloidin To test if mutant cochleae contained supporting cells we co-activator for all Notch molecules and thus deletion of this protein inhibits all Notch signaling. examined expression of supporting cell markers such as p27 and Prox1. On E17.5 p27 was expressed in supporting cells Methods and Measures ***** ***** under or between hair cells such as inner pharyngeal cells, Floxed Rbpsuh mice were crossed with Foxg1-Cre knock-in mice to delete the Rbpsuh gene in the inner ear. Inner ear phenotypes in Rbpsuh conditional knockout Deiter's cells and pillar cells (asterisks in figure 4 A and B). mice were determined at various developmental stages using immunohistochemistry. But no p27 expression was detected in those regions of RBP-J E Prox1 F Phalloidin G Prox1 H Phalloidin conditional knockout mice (Figure 4 C and D). -

C/Ebpα Is an Essential Collaborator in Hoxa9/Meis1-Mediated Leukemogenesis
C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis Cailin Collinsa, Jingya Wanga, Hongzhi Miaoa, Joel Bronsteina, Humaira Nawera, Tao Xua, Maria Figueroaa, Andrew G. Munteana, and Jay L. Hessa,b,1 aDepartment of Pathology, University of Michigan, Ann Arbor, MI 48109; and bIndiana University School of Medicine, Indianapolis, IN 46202 Edited* by Louis M. Staudt, National Institutes of Health, Bethesda, MD, and approved May 19, 2014 (received for review February 12, 2014) Homeobox A9 (HOXA9) is a homeodomain-containing transcrip- with Hoxa9. In addition, C/EBP recognition motifs are enriched tion factor that plays a key role in hematopoietic stem cell expan- at Hoxa9 binding sites. sion and is commonly deregulated in human acute leukemias. A C/EBPα is a basic leucine-zipper transcription factor that plays variety of upstream genetic alterations in acute myeloid leukemia a critical role in lineage commitment during hematopoietic dif- −/− (AML) lead to overexpression of HOXA9, almost always in associ- ferentiation (18). Whereas Cebpa mice show complete loss of ation with overexpression of its cofactor meis homeobox 1 (MEIS1). the granulocytic compartment, recent work shows that loss of α A wide range of data suggests that HOXA9 and MEIS1 play a syn- C/EBP in adult HSCs leads to both an increase in the number ergistic causative role in AML, although the molecular mechanisms of functional HSCs and an increase in their proliferative and leading to transformation by HOXA9 and MEIS1 remain elusive. In repopulating capacity (19, 20). Conversely, CEBPA overexpression can promote transdifferentiation of a variety of fibroblastic cells to this study, we identify CCAAT/enhancer binding protein alpha (C/ the myeloid lineage and can induce monocytic differentiation in EBPα) as a critical collaborator required for Hoxa9/Meis1-mediated α MLL-fusion protein-mediated leukemias (21, 22). -

The Suppressive Effects of 1,25-Dihydroxyvitamin D3 and Vitamin D Receptor on Brown Adipocyte Differentiation and Mitochondrial Respiration
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Masters Theses Graduate School 8-2014 The Suppressive Effects of 1,25-dihydroxyvitamin D3 and Vitamin D Receptor on Brown Adipocyte Differentiation and Mitochondrial Respiration Carolyn Jeanne Ricciardi University of Tennessee - Knoxville, [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_gradthes Part of the Molecular, Genetic, and Biochemical Nutrition Commons Recommended Citation Ricciardi, Carolyn Jeanne, "The Suppressive Effects of 1,25-dihydroxyvitamin D3 and Vitamin D Receptor on Brown Adipocyte Differentiation and Mitochondrial Respiration. " Master's Thesis, University of Tennessee, 2014. https://trace.tennessee.edu/utk_gradthes/2876 This Thesis is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Masters Theses by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a thesis written by Carolyn Jeanne Ricciardi entitled "The Suppressive Effects of 1,25-dihydroxyvitamin D3 and Vitamin D Receptor on Brown Adipocyte Differentiation and Mitochondrial Respiration." I have examined the final electronic copy of this thesis for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Master of Science, with a major in Nutrition. -

Test Summary Flyer-NGS Panels.Pub
Cancer‐related Mutaon Analysis Next Generaon Gene Sequencing for Myeloid Tesng Assay Summary IU Health Molecular Pathology Laboratory now offers high throughput sequencing for hot spot mutations found in clinically relevant cancer genes. In addition to a general panel of 54 genes, selected panels have been developed for a more tai- lored application in specific cancers. Comparing to single gene assay, these panels offer a more comprehensive and eco- nomic way to assess prognosis and/or treatment options for cancer patients at the initial diagnosis or at the relapse. Orderable Name: Use IU Health Molecular Pathology requisition; Call 317.491.6417 for requisition. Panels include: AML Mutations by NGS ASXL1, CEBPA, DNMT3A, ETV6/TEL, FLT3, HRAS, IDH1, IDH2, KIT, KRAS, MLL, NPM1, NRAS, PHF6, RUNX1, TET2, TP53, WT1 MDS Mutations by NGS ASXL1, ATRX, BCOR, BCORL1, ETV6/TEL, DNMT3A, EZH2, GNAS, IDH1, IDH2, RUNX1, SF3B1, SRSF2, TET2, TP53, U2AF1, ZRSR2 CML Mutations by NGS ABL1 MPN Mutations by NGS ASXL1, BRAF, CALR, CSF3R, EZH2, IKZF1, JAK2, JAK3, KDM6A, KIT, MPL, PDGRA, SETBP1, TET2 CMML Mutations by NGS ASXL1, CBL, CBLB, CBLC, EZH2, RUNX1, TET2, TP53, SRSF2 JMML Mutations by NGS CBL, CBLB, CBLC, HRAS, KRAS, NRAS, PTPN11 ALL Mutations by NGS ABL1, CSF3R, FBXW7, IKZF1, JAK3, KDM6A, NOTCH1 CLL Mutations by NGS MYD88, NOTCH1, SF3B1, TP53 Lymphoma/Myeloma Mutations by NGS BRAF, CDKN2A, CSF3R, FBXW7, HRAS, KRAS, MYD88, NOTCH1, NRAS, SF3B1,TP53 Hematopoietic Neoplasms Mutations by NGS ABL1, ASXL1, ATRX, BCOR, BCORL1, BRAF, CALR, CBL, CBLB, CBLC,CDKN2A, CEBPA, CSF3R, CUX1, DNMT3A, ETV6/TEL, EZH2, FBXW7, FLT3, GATA1, GATA2, GNAS, HRAS, IDH1, IDH2, IKZF1,JAK2, JAK3, KDM6A, KIT, KRAS, MLL, MPL, MYD88, NOTCH1, NPM1, NRAS, PDGFRA, PHF6, PTEN, PTPN11, RAD21, RUNX1,SETBP1, SF3B1, SMC1A, SMC3, SRSF2, STAG2, TET2, TP53, U2AF1, WT1, ZRSR2 Clinical Utility: This test is useful for the assessment of prognosis and/or treatment options for cancer patients at the initial diagnosis or at the relapse. -

Downloaded from Bioscientifica.Com at 09/26/2021 08:38:14PM Via Free Access
1 185 L C Gregory, P Gergics, OTX2 mutations in congenital 185:1 121–135 Clinical Study M Nakaguma and others hypopituitarism The phenotypic spectrum associated with OTX2 mutations in humans Louise C Gregory 1,*, Peter Gergics2,* , Marilena Nakaguma3,* , Hironori Bando2 , Giuseppa Patti1,4,5, Mark J McCabe1, Qing Fang 2, Qianyi Ma2 , Ayse Bilge Ozel2 , Jun Z Li2 , Michele Moreira Poina3, Alexander A L Jorge 3, Anna F Figueredo Benedetti3, Antonio M Lerario3, Ivo J P Arnhold3 , Berenice B Mendonca3 , Mohamad Maghnie4,5, Sally A Camper2 , Luciani R S Carvalho3 and Mehul T Dattani1 1Section of Molecular Basis of Rare Disease, Genetics and Genomic Medicine Research & Teaching Department, UCL Great Ormond Street Institute of Child Health, London, UK, 2Department of Human Genetics, University of Michigan, Correspondence Ann Arbor, Michigan, USA, 3Developmental Endocrinology Unit, Hospital das Clinicas da Faculdade de Medicina da should be addressed Universidade de São Paulo, São Paulo, Brazil, 4Department of Pediatrics, IRCCS Istituto Giannina Gaslini and to S A Camper or M T 5Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health, University of Dattani Genova, Genova, Italy Email *(L C Gregory, P Gergics and M Nakaguma contributed equally to this work) [email protected] or [email protected] Abstract Objective: The transcription factor OTX2 is implicated in ocular, craniofacial, and pituitary development. Design: We aimed to establish the contribution of OTX2 mutations in congenital hypopituitarism patients with/without eye abnormalities, study functional consequences, and establish OTX2 expression in the human brain, with a view to investigate the mechanism of action. Methods: We screened patients from the UK (n = 103), international centres (n = 24), and Brazil (n = 282); 145 were within the septo-optic dysplasia spectrum, and 264 had no eye phenotype. -

Nrf1 and Nrf2 Positively and C-Fos and Fra1 Negatively Regulate the Human Antioxidant Response Element-Mediated Expression of NAD(P)H:Quinone Oxidoreductase1 Gene
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 14960–14965, December 1996 Pharmacology Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene RADJENDIRANE VENUGOPAL AND ANIL K. JAISWAL* Department of Pharmacology, Fox Chase Cancer Center, 7701 Burholme Avenue, Philadelphia, PA 19111 Communicated by Y. W. Kan, University of California, San Francisco, CA, August 5, 1996 (received for review February 21, 1996) ABSTRACT Twenty-four base pairs of the human anti- (c-Jun, Jun-D, and Jun-B) form homodimers among them- oxidant response element (hARE) are required for high basal selves and heterodimers with c-Fos and Fra (Fra1 and Fra2) transcription of the NAD(P)H:quinone oxidoreductase1 proteins and bind to the TRE (a single AP1 binding site) and (NQO1) gene and its induction in response to xenobiotics and regulate the expression of target genes (6). Nrf1 and Nrf2 are antioxidants. hARE is a unique cis-element that contains one recently identified NF-E2-related factors that bind to the perfect and one imperfect AP1 element arranged as inverse sequence GCTGAGTCATGATGAGTCA (7, 8). The Nrf1 repeats separated by 3 bp, followed by a ‘‘GC’’ box. We report and Nrf2 contain 742 and 589 amino acids, respectively. Both here that Jun, Fos, Fra, and Nrf nuclear transcription factors these proteins display marked homology to the NF-E2, which bind to the hARE. Overexpression of cDNA derived combi- regulates expression of globin genes in developing erythroid nations of the nuclear proteins Jun and Fos or Jun and Fra1 cells through interaction with upstream AP1-like recognition repressed hARE-mediated chloramphenicol acetyltransferase sites (9). -

Genetic Testing for Acute Myeloid Leukemia AHS-M2062
Corporate Medical Policy Genetic Testing for Acute Myeloid Leukemia AHS-M2062 File Name: genetic_testing_for_acute_myeloid_leukemia Origination: 1/1/2019 Last CAP Review: 8/2021 Next CAP Review: 8/2022 Last Review: 8/2021 Description of Procedure or Service Acute myeloid leukemia (AML) is characterized by large numbers of abnormal, immature myeloid cells in the bone marrow and peripheral blood resulting from genetic changes in hematopoietic precursor cells which disrupt normal hematopoietic growth and differentiation (Stock, 2020). Related Policies: Genetic Cancer Susceptibility Using Next Generation Sequencing AHS-M2066 Molecular Panel Testing of Cancers to Identify Targeted Therapy AHS-M2109 Serum Tumor Markers for Malignancies AHS-G2124 Minimal Residual Disease (MRD) AHS- M2175 ***Note: This Medical Policy is complex and technical. For questions concerning the technical language and/or specific clinical indications for its use, please consult your physician. Policy BCBSNC will provide coverage for genetic testing for acute myeloid leukemia when it is determined to be medically necessary because the medical criteria and guidelines shown below are met. Benefits Application This medical policy relates only to the services or supplies described herein. Please refer to the Member's Benefit Booklet for availability of benefits. Member's benefits may vary according to benefit design; therefore member benefit language should be reviewed before applying the terms of this medical policy. When Genetic Testing for Acute Myeloid Leukemia is covered The use of genetic testing for acute myeloid leukemia is considered medically necessary for the following: A. Genetic testing for FLT3 internal tandem duplication and tyrosine kinase domain mutations (ITD and TKD), IDH1, IDH2, TET2, WT1, DNMT3A, ASXL1 and/or TP53 in adult and pediatric patients with suspected or confirmed AML of any type for prognostic and/or therapeutic purposes. -

Regulation of the C/Ebpα Signaling Pathway in Acute Myeloid Leukemia (Review)
ONCOLOGY REPORTS 33: 2099-2106, 2015 Regulation of the C/EBPα signaling pathway in acute myeloid leukemia (Review) GUANHUA SONG1, LIn Wang2, Kehong BI3 and guosheng JIang1 1Department of hemato-oncology, Institute of Basic Medicine, shandong academy of Medical sciences, Key Laboratory for Modern Medicine and Technology of Shandong Province, Key Laboratory for Rare and Uncommon Diseases, Key Medical Laboratory for Tumor Immunology and Traditional Chinese Medicine Immunology of shandong Province, Jinan, Shandong 250062; 2Research Center for Medical Biotechnology, Shandong Academy of Medical Sciences, Jinan, Shandong 250062; 3Department of Hematology, Qianfoshan Mountain Hospital of Shandong University, Jinan, Shandong 250014, P.R. China Received December 2, 2014; Accepted January 26, 2015 DoI: 10.3892/or.2015.3848 Abstract. The transcription factor CCAAT/enhancer binding Contents protein α (C/EBPα), as a critical regulator of myeloid devel- opment, directs granulocyte and monocyte differentiation. 1. Introduction Various mechanisms have been identified to explain how 2. Function of C/EBPα in myeloid differentiation C/EBPα functions in patients with acute myeloid leukemia 3. Regulation of the C/EBPα signaling pathway (AML). C/EBPα expression is suppressed as a result of 4. Conclusion common leukemia-associated genetic and epigenetic altera- tions such as AML1-ETO, RARα-PLZF or gene promoter methylation. Recent data have shown that ubiquitination modi- 1. Introduction fication also contributes to its downregulation. In addition, 10-15% of patients with AML in an intermediate cytogenetic Acute myeloid leukemia (AML) is characterized by uncon- risk subgroup were characterized by mutations of the C/EBPα trolled proliferation of myeloid progenitors that exhibit a gene.