Genetic Structure in Village Dogs Reveals a Central Asian Domestication Origin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Schedule Is Subject to Change Carolina Dog Judges Study Group
Schedule is Subject to Change Carolina Dog Judges Study Group Hound Breeds - Seminars and Workshops Registration TD Convention Center; 1 Exposition Avenue; Greenville, SC – July 2021 NAME_______________________________________ Judges #___________ ADDRESS_______________________________________________________ CITY_________________________ STATE _______ ZIP CODE ___________ Email Phone Please circle each breed that you expect to attend. This will help our presenters ensure that they have the proper amount of handout materials available. Schedule is subject to changes/adjustments. Thursday, July 29, 2021 9:00 to 11:45 am Basset Hound – presented by Kitty Steidel 12:00 to 2:30 pm Grand Bassets Griffons Vendeen - presented by Kitty Steidel 2:30 to 5:00 pm Petit Basset Griffon Vendeen - presented by Kitty Steidel 5:00 to 7:30 pm Basset Fauve de Bretagnes by Cindy Hartman Friday, July 30, 2021 9:00 to 11:30 am Cirneco dell’Etna – presented by Lucia Prieto 12:00 to 2:30 pm Greyhound presented by Patty Clark 2:30 to 5:00 pm Pharaoh Hound presented by Sheila Hoffman 5:00 – 7:30 pm Harrier presented by Kevin Shupenia Saturday, July 31, 2021 8:00 to 10:30 am Whippet – presented by Gail Boyd and Suzie Hughes 10:30 to 1:30 pm Borzoi - presented by Patti Neale 2:00 to 4:30 pm Scottish Deerhound -presented by Lynn Kiaer 4:30 to 7:00 pm Azawakh – presented by Fabian Arienti Sunday, August 1, 2021 8:30 to 11:00 am Basenji presented by Marianne Klinkowski 12:00 to 2:30 pm English Foxhound presented by Kevin Shupenia 2:30 to 5:00 pm American Foxhound presented by Polly Smith and Lisa Miller Return your completed application along with the payment of $60.00 per day ($75 per day payable after July 1, 2021) to: Cindy Stansell - 2199 Government Road, Clayton, NC 27520. -

Chrti Psi Bez Srsti
OBSAH Pošťáci 81 Eurasier 151 Psí burlaci 82 Šiperka 153 PE S 11 Závody saňových psů 82 Sibiřský husky 84 CHRTI Potom ek vlka 12 Aljašský malamut 88 Psí geny říkají něco jiného, než psí Sam ojed 90 Chrti východní, chrti západní 156 kosti 13 Grónský pes 93 Princip ucha 156 Genom psa 15 Kanadský inuitský pes 95 Tajemství skalního města 160 Vznik plemen a jejich Americký eskymácký pes 96 Šakalové, hyeny, chrti a bozi 161 klasifikace 18 Alaskáni a spol 96 Psi faraónů a „faraónský chrt" 164 První systematická nomenklatura Alaskan husky 97 Tesem 166 psů 18 C h in o o k 97 Faraónský chrt 166 Genetické skupiny 20 Evropský saňový pes 99 Domorodí chrti Indie 168 Plem ena 20 Z historie saňových psů Rampúrský chrt 168 v Čechách 100 Rajapalayam (Poligar) 169 DOMESTIKACE 21 S Byrdem na Jižní točnu 100 Čippiparai 169 Český horský pes 104 Pašm i 169 Když vlk sklopil uši 21 Kuči 169 Kdo si koho ochočil 21 Severští lovečtí špicové 107 Banjara 169 Domestikace 22 Norský losí pes šedý 110 Venuše a psi 24 Norský losí pes černý 110 Chrti Afriky a Asie 170 „Bez psa by člověk zůstal opicí" 24 Švédský losí pes 112 Afghánský chrt 170 Cesty psů jsou cestami lidí 28 Švédský bílý elkhound 112 Saluki 173 Pes v Evropě a zemědělství 29 Hälleforshund 112 Sloughi 175 Nejstarší historie psa v datech 32 Norský lundehund 113 Azavak 178 Zařazení plemen podle nomenkla Lajka ruskoevropská 115 Barzoj 180 tury FCI 34 Lajka západosibiřská 115 Chortaja borzaja 183 Lajka východosibřská 115 Tazi 185 SENSI PSI a PÁRIOVÉ 39 Lajka ruskofinská 115 Tajgan 186 Karelský medvědí pes 118 -

Secretary's Pages
SECRETARY ’S PAGES MISSION STATEMENT The American Kennel Club is dedicated to upholding the T ATENTION DELEGATES integrity of its RMegIisStrSy, IpOroNmo ting the spSorTt Aof TpEurMebrEeNd dT ogs and breeding for type and function. ® NOTICE OF MEETING TFohuen Admederiin ca1n8 8K4e, ntnhelAKC Cluba nisd dites daicffailtieadte td o ourpghaonlidziantgio nths ea idnvteogcarittey foofr iths e Rpeugriset brrye, dp rdoomgo atisn ga tfhame islyp ocrot mofpapnuiroenb,r ead vdaongcs e acnad nibnree ehdeianlgthf oarndty pwe elal-nbd eifnugn,c wtioonrk. to protect the rights of all Fdougn odwedneinrs1 a8n8d4 ,ptrhoe mAKCote raensd piotns saifbflieli adtoegd orwgnaenrizsahtipio. ns advocate for the pure bred dog as a The next meeting of the Delegates will be held family companion, advance canine health and well-being, work to protect the rights of all dog owners and 805prom1 oAtrec ore Csopropnosribaltee dDorgiv oew, Snueirtseh 1ip0. 0, Raleigh, NC 276 17 at the Doubletree Newark Airport Hotel on 101 Park Avenue, New York, NY 10178 8051 Arco Corporate Drive, Suite 100, Raleigh, NC 276 17 Tuesday Raleigh, NC Customer Call Center ..............................................................(919) 233-9767 260 Madison Avenue, New York, NY 10016 , September 10, 2019. For the sole pur- New York, NY Office ...................................................................................(212) 696-8200 Raleigh, NC Customer Call Center ..............................................................(919) 233-9767 Fax .............................................................................................................(212) -

Dog Breeds of the World
Dog Breeds of the World Get your own copy of this book Visit: www.plexidors.com Call: 800-283-8045 Written by: Maria Sadowski PlexiDor Performance Pet Doors 4523 30th St West #E502 Bradenton, FL 34207 http://www.plexidors.com Dog Breeds of the World is written by Maria Sadowski Copyright @2015 by PlexiDor Performance Pet Doors Published in the United States of America August 2015 All rights reserved. No portion of this book may be reproduced or transmitted in any form or by any electronic or mechanical means, including photocopying, recording, or by any information retrieval and storage system without permission from PlexiDor Performance Pet Doors. Stock images from canstockphoto.com, istockphoto.com, and dreamstime.com Dog Breeds of the World It isn’t possible to put an exact number on the Does breed matter? dog breeds of the world, because many varieties can be recognized by one breed registration The breed matters to a certain extent. Many group but not by another. The World Canine people believe that dog breeds mostly have an Organization is the largest internationally impact on the outside of the dog, but through the accepted registry of dog breeds, and they have ages breeds have been created based on wanted more than 340 breeds. behaviors such as hunting and herding. Dog breeds aren’t scientifical classifications; they’re It is important to pick a dog that fits the family’s groupings based on similar characteristics of lifestyle. If you want a dog with a special look but appearance and behavior. Some breeds have the breed characterics seem difficult to handle you existed for thousands of years, and others are fairly might want to look for a mixed breed dog. -

BCOA Bulleint May-June-July-August 2017
Vol. 52 | No. 2 | MAY JUN JUL AUG | 2017 Th e Offi cial PublicationBULLETIN of the Basenji Club of America, Inc. cvr2 BCOA Bulletin (MAY/JUN/JUL/AUG 2017) visit us online at www.basenji.org www.facebook.com/basenji.org BCOA Bulletin (MAY/JUN/JUL/AUG 2017) 1 2 BCOA Bulletin (MAY/JUN/JUL/AUG 2017) visit us online at www.basenji.org www.facebook.com/basenji.org BCOA Bulletin (MAY/JUN/JUL/AUG 2017) 3 BCOA BULLETIN Tootsie’s get is as follows in the order they fi nished: CONTENTS MAY/JUN/JUL/AUG 2017 1. BISS Ch. Taji’s Klassic Beauty – #1 bitch in 2005, AOM and Best Veteran at national 2. Am/Int. UKC Ch. Klassic’s Hot Ticket to Berimo – WB 2004 national at 9 mos. Old. 3. Am/Eng. MBISS/MBIS Klassic’s Million Dollar Baby at Tokaji – Millie has broken every record in the UK, won Cruft s best of breed 5 times, Top Basenji, Top Hound and 2 times Top Brood On the cover bitch all breeds. 4. DC Taji’s Klassic Architecture SC SDHR – Finished at 8 mos of age at the EBC specialty, Tootsie WWWHA grand sweep winner of over 100 hounds. MBIS/BISS CDN/MBISS CH. KLASSIC’S ROOT TOOT TOOT 5. Ch. Klassic’s Hot to Trot to Naharin – fi nished with all majors and lives on the couch in CA. 6. DC Klassic’s Ms Mata Hauri SC – best in sweeps at the 2005 national, BOB at the 2006 national, 7 time BIS winner (holds record for top bitch), 2 time BOB winner Westminster KC – lives in Who doesn’t know the name Tootsie??? Tootsie had a wonderful show career – NH on Debbie’s couch. -

The Intelligence of Dogs a Guide to the Thoughts, Emotions, and Inner Lives of Our Canine
Praise for The Intelligence of Dogs "For those who take the dog days literally, the best in pooch lit is Stanley Coren’s The Intelligence of Dogs. Psychologist, dog trainer, and all-around canine booster, Coren trots out everyone from Aristotle to Darwin to substantiate the smarts of canines, then lists some 40 commands most dogs can learn, along with tests to determine if your hairball is Harvard material.” —U.S. News & World Report "Fascinating . What makes The Intelligence of Dogs such a great book, however, isn’t just the abstract discussions of canine intelli gence. Throughout, Coren relates his findings to the concrete, dis cussing the strengths and weaknesses of various breeds and including specific advice on evaluating different breeds for vari ous purposes. It's the kind of book would-be dog owners should be required to read before even contemplating buying a dog.” —The Washington Post Book World “Excellent book . Many of us want to think our dog’s persona is characterized by an austere veneer, a streak of intelligence, and a fearless-go-for-broke posture. No matter wrhat your breed, The In telligence of Dogs . will tweak your fierce, partisan spirit . Coren doesn’t stop at intelligence and obedience rankings, he also explores breeds best suited as watchdogs and guard dogs . [and] does a masterful job of exploring his subject's origins, vari ous forms of intelligence gleaned from genetics and owner/trainer conditioning, and painting an inner portrait of the species.” —The Seattle Times "This book offers more than its w7ell-publicized ranking of pure bred dogs by obedience and working intelligence. -
Domestic Dog Breeding Has Been Practiced for Centuries Across the a History of Dog Breeding Entire Globe
ANCESTRY GREY WOLF TAYMYR WOLF OF THE DOMESTIC DOG: Domestic dog breeding has been practiced for centuries across the A history of dog breeding entire globe. Ancestor wolves, primarily the Grey Wolf and Taymyr Wolf, evolved, migrated, and bred into local breeds specific to areas from ancient wolves to of certain countries. Local breeds, differentiated by the process of evolution an migration with little human intervention, bred into basal present pedigrees breeds. Humans then began to focus these breeds into specified BREED Basal breed, no further breeding Relation by selective Relation by selective BREED Basal breed, additional breeding pedigrees, and over time, became the modern breeds you see Direct Relation breeding breeding through BREED Alive migration BREED Subsequent breed, no further breeding Additional Relation BREED Extinct Relation by Migration BREED Subsequent breed, additional breeding around the world today. This ancestral tree charts the structure from wolf to modern breeds showing overlapping connections between Asia Australia Africa Eurasia Europe North America Central/ South Source: www.pbs.org America evolution, wolf migration, and peoples’ migration. WOLVES & CANIDS ANCIENT BREEDS BASAL BREEDS MODERN BREEDS Predate history 3000-1000 BC 1-1900 AD 1901-PRESENT S G O D N A I L A R T S U A L KELPIE Source: sciencemag.org A C Many iterations of dingo-type dogs have been found in the aborigine cave paintings of Australia. However, many O of the uniquely Australian breeds were created by the L migration of European dogs by way of their owners. STUMPY TAIL CATTLE DOG Because of this, many Australian dogs are more closely related to European breeds than any original Australian breeds. -

The Evolutionary History of Dogs in the Americas Authors
1 Title: The Evolutionary History of Dogs in the Americas 2 3 Authors: Máire Ní Leathlobhair1*, Angela R. Perri2,3*, Evan K. Irving-Pease4*, Kelsey E. Witt5*, 4 Anna Linderholm4,6*, James Haile4,7, Ophelie Lebrasseur4, Carly Ameen8, Jeffrey Blick9,†, Adam R. 5 Boyko10, Selina Brace11, Yahaira Nunes Cortes12, Susan J. Crockford13, Alison Devault14, 6 Evangelos A. Dimopoulos4, Morley Eldridge15, Jacob Enk14, Shyam Gopalakrishnan7, Kevin Gori1, 7 Vaughan Grimes16, Eric Guiry17 , Anders J. Hansen7,18, Ardern Hulme-Beaman4,8, John Johnson19, 8 Andrew Kitchen20, Aleksei K. Kasparov21, Young-Mi Kwon1, Pavel A. Nikolskiy21,22, Carlos 9 Peraza Lope23, Aurélie Manin24,25, Terrance Martin26, Michael Meyer27, Kelsey Noack Myers28, 10 Mark Omura29, Jean-Marie Rouillard14,30, Elena Y. Pavlova21,31, Paul Sciulli32, Mikkel-Holger S. 11 Sinding7,18,33, Andrea Strakova1, Varvara V. Ivanova34 , Christopher Widga35, Eske Willerslev7, 12 Vladimir V. Pitulko21, Ian Barnes11, M. Thomas P. Gilbert7,36, Keith M. Dobney8, Ripan S. 13 Malhi37,38, Elizabeth P. Murchison1,a,§, Greger Larson4,a,§ and Laurent A. F. Frantz4,39,a,§ 14 15 Affiliations: 16 1 Transmissible Cancer Group, Department of Veterinary Medicine, University of Cambridge, 17 Cambridge, U.K. 18 2 Department of Archaeology, Durham University, Durham, U.K. 19 3 Department of Human Evolution, Max Planck Institute for Evolutionary Anthropology, Leipzig, 20 Germany 21 4 The Palaeogenomics & Bio-Archaeology Research Network, Research Laboratory for 22 Archaeology and History of Art, The University of Oxford, -
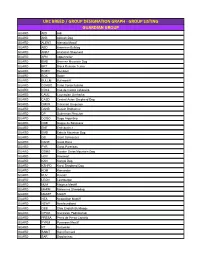
Ukc Breed / Group Designation Graph
UKC BREED / GROUP DESIGNATION GRAPH - GROUP LISTING GUARDIAN GROUP GUARD AIDI Aidi GUARD AKB Akbash Dog GUARD ALENT Alentejo Mastiff GUARD ABD American Bulldog GUARD ANAT Anatolian Shepherd GUARD APN Appenzeller GUARD BMD Bernese Mountain Dog GUARD BRT Black Russian Terrier GUARD BOER Boerboel GUARD BOX Boxer GUARD BULLM Bullmastiff GUARD CORSO Cane Corso Italiano GUARD CDCL Cao de Castro Laboreiro GUARD CAUC Caucasian Ovcharka GUARD CASD Central Asian Shepherd Dog GUARD CMUR Cimarron Uruguayo GUARD DANB Danish Broholmer GUARD DP Doberman Pinscher GUARD DOGO Dogo Argentino GUARD DDB Dogue de Bordeaux GUARD ENT Entlebucher GUARD EMD Estrela Mountain Dog GUARD GS Giant Schnauzer GUARD DANE Great Dane GUARD PYR Great Pyrenees GUARD GSMD Greater Swiss Mountain Dog GUARD HOV Hovawart GUARD KAN Kangal Dog GUARD KSHPD Karst Shepherd Dog GUARD KOM Komondor GUARD KUV Kuvasz GUARD LEON Leonberger GUARD MJM Majorca Mastiff GUARD MARM Maremma Sheepdog GUARD MASTF Mastiff GUARD NEA Neapolitan Mastiff GUARD NEWF Newfoundland GUARD OEB Olde English Bulldogge GUARD OPOD Owczarek Podhalanski GUARD PRESA Perro de Presa Canario GUARD PYRM Pyrenean Mastiff GUARD RT Rottweiler GUARD SAINT Saint Bernard GUARD SAR Sarplaninac GUARD SC Slovak Cuvac GUARD SMAST Spanish Mastiff GUARD SSCH Standard Schnauzer GUARD TM Tibetan Mastiff GUARD TJAK Tornjak GUARD TOSA Tosa Ken SCENTHOUND GROUP SCENT AD Alpine Dachsbracke SCENT B&T American Black & Tan Coonhound SCENT AF American Foxhound SCENT ALH American Leopard Hound SCENT AFVP Anglo-Francais de Petite Venerie SCENT -

United Greyhound Club Conformation Entries 8:30 – 9:30 Am Show Starts 10:00 Am
Weekend Schedule Saturday United Greyhound Club Conformation entries 8:30 – 9:30 am Show starts 10:00 am Lure Coursing Entries 11:00 am – 12:00 noon Handler's meeting and draw 12:00 noon LURE COURSING 2 Trials, 4 CA Tests Sunday Lure Coursing Entries 9:90 – 10:00 am Saturday and Sunday, Sept 5 & 6, 2020 Handler's meeting and draw 10:00 am CONFORMATION Sighthound Specialty Directions from Toronto: Take ON-401 to Norwich Ave/Oxford 59 in Woodstock. Take Laliberty's Farm Exit 232 from ON-401 W 1174 Norfolk Cty Rd 21 Follow Oxford 59, Oxford Rd 13/Regional Rd 13 and Norfolk Delhi, ON N4B 2W4 County HW59 to Norfolk County Road 21 Turn left on Norfolk County Road 21 (Andy’s Drive-Inn Snack Pre- entries close bar is at the corner of HW 59/Norfolk County Rd21) Wed, Aug 26, 2020 at 8 pm Drive 1.7km and your destination will be on your right. Follow the sign for the entrance on the field. COVID-19 Restrictions All Ontario and local Health Unit requirements will be observed. All participants must wear a mask when in the ring, and when slipping dogs in the field trial. Masks will be available for a small donation, if needed. Please allow space between cars when parking. Please clean up after your dogs and take your own garbage home with you. The show ring will have separate entrance and exits, and ribbons will be self-serve as you leave the ring. LURE COURSING Use the attached entry form, or use the fillable ones on the Judge: All three days: Katie Stoyles, Newmarket, ON UKC site. -

PREMIUM LISTS SATURDAY May 24, 2016
This is the skeleton Premium for all shows. It has everything except the Show Name, Show Location & Judges. This information will be linked on the show you have selected. PREMIUM LISTS Hosted by, the American Rare Breed Association Example SATURDAY May 24, 2016 The American Rare Breed Association and its sister organization the Kennel Club USA staff do not enter dogs into our conformation dog show events. SHOW HOURS – 7: OO am to 6:00 pm Example Motel 6 840 S. Indian Hill Blvd. Claremont California 91711 NATIONAL & INTERNATIONAL CHAMPIONSHIPS SPAYED & NEUTERED CLASSES This is an outdoor venue. You can bring along EZ-Up’s to provide shade for you and your dogs. This show is located on the grounds of the Motel 6, call Steven to make your reservations. He can be reached at 909-621- 4831. Be sure to mention that you are showing your dog with the American Rare Breed Association. This show will be co-hosted by Kennel Club-USA. Kennel Club USA offers conformation shows for both National and International championships. Please call them to register your dog. They can be reached at 301- 868-8284. American Rare Breed Association 9921 Frank Tippett Road Cheltenham, MD 20623 Telephone: 301-868-5718 – Fax: 301-868-6409 http://www.arba.org sales @ arba.org http://www.arba.org/rules_regulations.htm FEES AND AWARDS PRE-ENTRY ENTRY FEES: The following fees are for each dog for each show. Shows 1 thru 6 3 to 6 month dogs, if you are a member……………………………..…………………………….…$20.00 3 to 6 month dogs, Non-Member………………………………………………………………………….$23.00 6 months or older if you are a member…………………………………………………………………$23.00 6 months or older Non-Member………………………………,,………………………………………….$25.00 Your dog can be entered in all of the shows for the event; however the dog cannot be entered into less than 3 shows. -

Dog Breeds Volume 5
Dog Breeds - Volume 5 A Wikipedia Compilation by Michael A. Linton Contents 1 Russell Terrier 1 1.1 History ................................................. 1 1.1.1 Breed development in England and Australia ........................ 1 1.1.2 The Russell Terrier in the U.S.A. .............................. 2 1.1.3 More ............................................. 2 1.2 References ............................................... 2 1.3 External links ............................................. 3 2 Saarloos wolfdog 4 2.1 History ................................................. 4 2.2 Size and appearance .......................................... 4 2.3 See also ................................................ 4 2.4 References ............................................... 4 2.5 External links ............................................. 4 3 Sabueso Español 5 3.1 History ................................................ 5 3.2 Appearance .............................................. 5 3.3 Use .................................................. 7 3.4 Fictional Spanish Hounds ....................................... 8 3.5 References .............................................. 8 3.6 External links ............................................. 8 4 Saint-Usuge Spaniel 9 4.1 History ................................................. 9 4.2 Description .............................................. 9 4.2.1 Temperament ......................................... 10 4.3 References ............................................... 10 4.4 External links