Comparative Analysis of Human Chromosome 7Q21 and Mouse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

G Protein-Coupled Receptors
www.aladdin-e.com Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: [email protected] Email EU: [email protected] Email Asia Pacific: [email protected] G PROTEIN-COUPLED RECEPTORS Overview: The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20–24 hydrophobic amino acids arranged as a-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Fredriksson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor (see Page S195) has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. -

Comparative Analysis of Human Chromosome 7Q21 and Mouse
Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing David Monk,1,6 Alexandre Wagschal,2 Philippe Arnaud,2 Pari-Sima Mu¨ller,3 Layla Parker-Katiraee,4 Déborah Bourc’his,5 Stephen W. Scherer,4 Robert Feil,2 Philip Stanier,1 and Gudrun E. Moore1 1Institute of Child Health, London WC1N 1EH, United Kingdom; 2Institute of Molecular Genetics, CNRS UMR-5535 and University of Montpellier-II, 34293 Montpellier, France; 3Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, United Kingdom; 4Center for Applied Genomics, The Hospital for Sick Children, Toronto M5G 1L7, Canada; 5Inserm U741, F-75251 Paris Cedex 05, France Genomic imprinting is a developmentally important mechanism that involves both differential DNA methylation and allelic histone modifications. Through detailed comparative characterization, a large imprinted domain mapping to chromosome 7q21 in humans and proximal chromosome 6 in mice was redefined. This domain is organized around a maternally methylated CpG island comprising the promoters of the adjacent PEG10 and SGCE imprinted genes. Examination of Dnmt3l−/+ conceptuses shows that imprinted expression for all genes of the cluster depends upon the germline methylation at this putative “imprinting control region” (ICR). Similarly as for other ICRs, we find its DNA-methylated allele to be associated with trimethylation of lysine 9 on histone H3 (H3K9me3) and trimethylation of lysine 20 on histone H4 (H4K20me3), whereas the transcriptionally active paternal allele is enriched in H3K4me2 and H3K9 acetylation. -

G Protein-Coupled Receptors
G PROTEIN-COUPLED RECEPTORS Overview:- The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20-24 hydrophobic amino acids arranged as α-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Frederikson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G-proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. Signalling through GPCR is enacted by the activation of heterotrimeric GTP-binding proteins (G proteins), made up of α, β and γ subunits, where the α and βγ subunits are responsible for signalling. The α subunit (tabulated below) allows definition of one series of signalling cascades and allows grouping of GPCRs to suggest common cellular, tissue and behavioural responses. -

Plasma Cells in Vitro Generation of Long-Lived Human
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology , 32 of which you can access for free at: 2012; 189:5773-5785; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication November 2012; doi: 10.4049/jimmunol.1103720 http://www.jimmunol.org/content/189/12/5773 In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco, Sophie Stephenson, Matthew A. Care, Darren Newton, Nicholas A. Barnes, Adam Davison, Andy Rawstron, David R. Westhead, Gina M. Doody and Reuben M. Tooze J Immunol cites 65 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/11/16/jimmunol.110372 0.DC1 This article http://www.jimmunol.org/content/189/12/5773.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. The Journal of Immunology In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco,*,1 Sophie Stephenson,*,1 Matthew A. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -
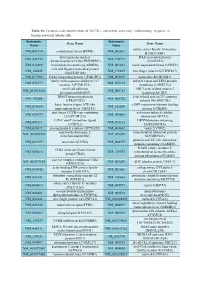
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

GNG11 (G-Protein Γ Subunit 11) Suppresses Cell Growth with Induction of Reactive
Biochemistry and Cell Biology GNG11 (G-protein γ subunit 11) suppresses cell growth with induction of reactive oxygen species and abnormal nuclear morphology in human SUSM1 cells Journal: Biochemistry and Cell Biology Manuscript ID bcb-2016-0248.R1 Manuscript Type: Article Date Submitted by the Author: 02-Mar-2017 Complete List of Authors: Takauji, Yuki; Yokohama City University Kudo, Ikuru; Yokohama City University En, Atsuki; DraftYokohama City University Matsuo, Ryo; Yokohama City University Hossain, Mohammad; Primeasia University Nakabayashi, Kazuhiko; National Center for Child Health and Development Miki, Kensuke; Yokohama City University Fujii, Michihiko; Yokohama City University Ayusawa, Dai; Yokohama City University Please Select from this Special N/A Issues list if applicable: GNG11, cellular senescence, cellular immortalization, nuclear shape, Keyword: reactive oxygen species https://mc06.manuscriptcentral.com/bcb-pubs Page 1 of 33 Biochemistry and Cell Biology 1 GNG11 (G-protein γ subunit 11) suppresses cell growth with induction of reactive 2 oxygen species and abnormal nuclear morphology in human SUSM-1 cells 3 4 Yuki Takauji1,5, Ikuru Kudo1,5, Atsuki En1, Ryo Matsuo1, Mohammad Nazir Hossain3, Kazuhiko 5 Nakabayashi4, Kensuke Miki1,2, Michihiko Fujii1* and Dai Ayusawa1,2 6 7 1Graduate school of Nanobioscience, Yokohama City University, 22-2 Seto, Kanazawa-ku, 8 Yokohama, Kanagawa 236-0027, JapanDraft 9 2Ichiban Life Corporation, 1-1-7 Horai-cho, Naka-ku, Yokohama, Kanagawa 231-0048, Japan 10 3Department of Biochemistry, Primeasia University, 9 Banani C/A Banani, Dhaka 1213, Bangladesh 11 4Department of Maternal-Fetal Biology, National Center for Child Health and Development, Tokyo 12 157-8535, Japan 13 5Both authors contributed equally to this work. -

De-Repression of the RAC Activator ELMO1 in Cancer Stem Cells
RESEARCH ARTICLE De-repression of the RAC activator ELMO1 in cancer stem cells drives progression of TGFb-deficient squamous cell carcinoma from transition zones Heather A McCauley1*, Ve´ ronique Chevrier2, Daniel Birnbaum2, Ge´ raldine Guasch1,2* 1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, United States; 2Centre de Recherche en Cance´rologie de Marseille (CRCM), Inserm, U1068, F-13009, CNRS, UMR7258, F-13009, Institut Paoli- Calmettes, F-13009, Aix-Marseille University, UM 105, F-13284, Marseille, France Abstract Squamous cell carcinomas occurring at transition zones are highly malignant tumors with poor prognosis. The identity of the cell population and the signaling pathways involved in the progression of transition zone squamous cell carcinoma are poorly understood, hence representing limited options for targeted therapies. Here, we identify a highly tumorigenic cancer stem cell population in a mouse model of transitional epithelial carcinoma and uncover a novel mechanism by which loss of TGFb receptor II (Tgfbr2) mediates invasion and metastasis through de-repression of ELMO1, a RAC-activating guanine exchange factor, specifically in cancer stem cells of transition zone tumors. We identify ELMO1 as a novel target of TGFb signaling and show that restoration of Tgfbr2 results in a complete block of ELMO1 in vivo. Knocking down Elmo1 impairs metastasis of carcinoma cells to the lung, thereby providing insights into the mechanisms of progression of *For correspondence: Heather. Tgfbr2-deficient invasive transition zone squamous cell carcinoma. [email protected] (HAM); DOI: 10.7554/eLife.22914.001 geraldine.guasch-grangeon@ inserm.fr (GG) Competing interests: The authors declare that no Introduction competing interests exist. -

Supplementary Figure 1 Standardization of Gene Expression
Supplementary Figure 1 Standardization of gene expression Notes: (A) Standardization of GSE86544, (B) standardization of GSE103479, (C) standardization of GSE102238, (D) Standardization of GSE7055. The blue bar represents the data before normalization, and the red bar represents the data after normalization. Supplementary Figure 2 Correlation between module eigengenes and clinical traits especially PNI in GSE103479 and GSE102238 datasets. Notes: (A, B) Module-trait relationships in GSE103479 and GSE102238 datasets. The correlation coefficients and corresponding P-values in the brackets are contained in each cell. The table is color- coded by correlation between eigengenes and traits according to the color legend on the right side. The modules with the most significant differences are displayed in brackets. Abbreviations: PNI, perineural invasion. Supplementary Figure 3 The expression values of CCNB2 in pancreatic cancer (GSE102238) and colon cancer (GSE103479). Notes: (A, B) CCNB2 expression values were detected in GSE102238 and GSE103479. Abbreviations: CCNB2, cyclin B2 Supplementary Table 1 Results of top 20 pathway enrichment analysis of GSE7055 Term Category Description Count Log10(P) Genes GO:0000280 GO Biological nuclear division 33 -23.4 BIRC5,BUB1B,CCNB1,CCNE1,CDC20, Processes CKS2,KIF11,MAD2L1,MYBL2,SPAST, TOP2A,TTK,PRC1,PKMYT1,PTTG1,T RIP13,DLGAP5,TACC3,SMC2,SPAG5, UBE2C,ZWINT,TPX2,FBXO5,RACGA P1,NUSAP1,SPDL1,CDCA8,CEP55,ND C1,NSFL1C,KIF18B,ASPM GO:1902850 GO Biological microtubule 15 -12.89 BIRC5,CCNB1,CDC20,KIF11,MAD2L1 Processes -

SUPPORTING INFORMATION for Regulation of Gene Expression By
SUPPORTING INFORMATION for Regulation of gene expression by the BLM helicase correlates with the presence of G4 motifs Giang Huong Nguyen1,2, Weiliang Tang3, Ana I. Robles1, Richard P. Beyer4, Lucas T. Gray5, Judith A. Welsh1, Aaron J. Schetter1, Kensuke Kumamoto1,6, Xin Wei Wang1, Ian D. Hickson2,7, Nancy Maizels5, 3,8 1 Raymond J. Monnat, Jr. and Curtis C. Harris 1Laboratory of Human Carcinogenesis, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, U.S.A; 2Department of Medical Oncology, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford, U.K.; 3Department of Pathology, University of Washington, Seattle, WA U.S.A.; 4 Center for Ecogenetics and Environmental Health, University of Washington, Seattle, WA U.S.A.; 5Department of Immunology and Department of Biochemistry, University of Washington, Seattle, WA U.S.A.; 6Department of Organ Regulatory Surgery, Fukushima Medical University, Fukushima, Japan; 7Cellular and Molecular Medicine, Nordea Center for Healthy Aging, University of Copenhagen, Denmark; 8Department of Genome Sciences, University of WA, Seattle, WA U.S.A. SI Index: Supporting Information for this manuscript includes the following 19 items. A more detailed Materials and Methods section is followed by 18 Tables and Figures in order of their appearance in the manuscript text: 1) SI Materials and Methods 2) Figure S1. Study design and experimental workflow. 3) Figure S2. Immunoblot verification of BLM depletion from human fibroblasts. 4) Figure S3. PCA of mRNA and miRNA expression in BLM-depleted human fibroblasts. 5) Figure S4. qPCR confirmation of mRNA array data. 6) Table S1. BS patient and control detail. -

SUPPLEMENTARY APPENDIX Exome Sequencing Reveals Heterogeneous Clonal Dynamics in Donor Cell Myeloid Neoplasms After Stem Cell Transplantation
SUPPLEMENTARY APPENDIX Exome sequencing reveals heterogeneous clonal dynamics in donor cell myeloid neoplasms after stem cell transplantation Julia Suárez-González, 1,2 Juan Carlos Triviño, 3 Guiomar Bautista, 4 José Antonio García-Marco, 4 Ángela Figuera, 5 Antonio Balas, 6 José Luis Vicario, 6 Francisco José Ortuño, 7 Raúl Teruel, 7 José María Álamo, 8 Diego Carbonell, 2,9 Cristina Andrés-Zayas, 1,2 Nieves Dorado, 2,9 Gabriela Rodríguez-Macías, 9 Mi Kwon, 2,9 José Luis Díez-Martín, 2,9,10 Carolina Martínez-Laperche 2,9* and Ismael Buño 1,2,9,11* on behalf of the Spanish Group for Hematopoietic Transplantation (GETH) 1Genomics Unit, Gregorio Marañón General University Hospital, Gregorio Marañón Health Research Institute (IiSGM), Madrid; 2Gregorio Marañón Health Research Institute (IiSGM), Madrid; 3Sistemas Genómicos, Valencia; 4Department of Hematology, Puerta de Hierro General University Hospital, Madrid; 5Department of Hematology, La Princesa University Hospital, Madrid; 6Department of Histocompatibility, Madrid Blood Centre, Madrid; 7Department of Hematology and Medical Oncology Unit, IMIB-Arrixaca, Morales Meseguer General University Hospital, Murcia; 8Centro Inmunológico de Alicante - CIALAB, Alicante; 9Department of Hematology, Gregorio Marañón General University Hospital, Madrid; 10 Department of Medicine, School of Medicine, Com - plutense University of Madrid, Madrid and 11 Department of Cell Biology, School of Medicine, Complutense University of Madrid, Madrid, Spain *CM-L and IB contributed equally as co-senior authors. Correspondence: