Evolution of Transducin Alpha, Beta and Gamma Subunit Gene Families
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -
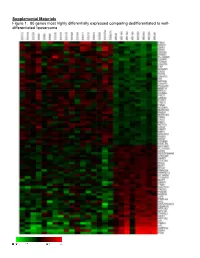
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

Identification of Residues of the H-Ras Protein Critical for Functional Interaction with Guanine Nucleotide Exchange Factors RAYMOND D
MOLECULAR AND CELLULAR BIOLOGY, Feb. 1994, p. 1104-1112 Vol. 14, No. 2 0270-7306/94/$04.00+0 Copyright © 1994, American Society for Microbiology Identification of Residues of the H-Ras Protein Critical for Functional Interaction with Guanine Nucleotide Exchange Factors RAYMOND D. MOSTELLER, JAEWON HAN, AND DANIEL BROEK* Department ofBiochemistry and Molecular Biology, Kenneth Norris Jr. Cancer Hospital and Research Center, University of Southern California School ofMedicine, Los Angeles, California 90033 Received 10 September 1993/Returned for modification 21 October 1993/Accepted 29 October 1993 Ras proteins are activated in vivo by guanine nucleotide exchange factors encoded by genes homologous to the CDC25 gene ofSaccharomyces cerevisiae. We have taken a combined genetic and biochemical approach to probe the sites on Ras proteins important for interaction with such exchange factors and to further probe the mechanism ofCDC25-catalyzed GDP-GTP exchange. Random mutagenesis coupled with genetic selection in S. cerevisiae was used to generate second-site mutations within human H-ras-alalS which could suppress the ability of the Ala-15 substitution to block CDC25 function. We transferred these second-site suppressor mutations to normal H-ras and oncogenic H-rasva1l2 to test whether they induced a general loss of function or whether they selectively affected CDC25 interaction. Four highly selective mutations were discovered, and they affected the surface-located amino acid residues 62, 63, 67, and 69. Two lines of evidence suggested that these residues may be involved in binding to CDC25: (i) using the yeast two-hybrid system, we demonstrated that these mutants cannot bind CDC25 under conditions where the wild-type H-Ras protein can; (ii) we demonstrated that the binding to H-Ras of monoclonal antibody Y13-259, whose epitope has been mapped to residues 63, 65, 66, 67, 70, and 73, is blocked by the mouse sosl and yeast CDC25 gene products. -

Investigating Cone Photoreceptor Development Using Patient-Derived NRL Null Retinal Organoids
ARTICLE https://doi.org/10.1038/s42003-020-0808-5 OPEN Investigating cone photoreceptor development using patient-derived NRL null retinal organoids Alyssa Kallman1,11, Elizabeth E. Capowski 2,11, Jie Wang 3, Aniruddha M. Kaushik4, Alex D. Jansen2, Kimberly L. Edwards2, Liben Chen4, Cynthia A. Berlinicke3, M. Joseph Phillips2,5, Eric A. Pierce6, Jiang Qian3, ✉ ✉ Tza-Huei Wang4,7, David M. Gamm2,5,8 & Donald J. Zack 1,3,9,10 1234567890():,; Photoreceptor loss is a leading cause of blindness, but mechanisms underlying photoreceptor degeneration are not well understood. Treatment strategies would benefit from improved understanding of gene-expression patterns directing photoreceptor development, as many genes are implicated in both development and degeneration. Neural retina leucine zipper (NRL) is critical for rod photoreceptor genesis and degeneration, with NRL mutations known to cause enhanced S-cone syndrome and retinitis pigmentosa. While murine Nrl loss has been characterized, studies of human NRL can identify important insights for human retinal development and disease. We utilized iPSC organoid models of retinal development to molecularly define developmental alterations in a human model of NRL loss. Consistent with the function of NRL in rod fate specification, human retinal organoids lacking NRL develop S- opsin dominant photoreceptor populations. We report generation of two distinct S-opsin expressing populations in NRL null retinal organoids and identify MEF2C as a candidate regulator of cone development. 1 Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, USA. 2 Waisman Center, University of Wisconsin-Madison, Madison, USA. 3 Department of Ophthalmology, Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, USA. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Calmodulin-Dependent Protein Kinase II–Protein Phosphatase 1 Switch Facilitates Specificity in Postsynaptic Calcium Signaling
An ultrasensitive Ca2؉͞calmodulin-dependent protein kinase II–protein phosphatase 1 switch facilitates specificity in postsynaptic calcium signaling J. Michael Bradshaw*†‡, Yoshi Kubota*, Tobias Meyer†, and Howard Schulman* Departments of *Neurobiology and †Molecular Pharmacology, Stanford University School of Medicine, Stanford, CA 94305 Edited by Roger A. Nicoll, University of California, San Francisco, CA, and approved July 21, 2003 (received for review May 7, 2003) The strength of hippocampal synapses can be persistently in- hence would provide a distinct Ca2ϩ activation region for creased by signals that activate Ca2؉͞calmodulin-dependent pro- CaMKII compared with other Ca2ϩ-activated enzymes at the tein kinase II (CaMKII). This CaMKII-dependent long-term potenti- synapse. In fact, it has been hypothesized that the signaling ation is important for hippocampal learning and memory. In this network controlling CaMKII autophosphorylation is a bistable work we show that CaMKII exhibits an intriguing switch-like type of switch that allows CaMKII to remain autophosphory- activation that likely is important for changes in synaptic strength. lated long after Ca2ϩ returns to a basal level (16). We found that autophosphorylation of CaMKII by itself showed a In this work we demonstrate experimentally that CaMKII 2؉ Ϸ ϩ steep dependence on Ca concentration [Hill coefficient (nH) 5]. responds in a switch-like fashion to Ca2 : CaMKII transitions 2؉ Ϸ However, an even steeper Ca dependence (nH 8) was observed rapidly from little to near-total autophosphorylation over a when autophosphorylation is balanced by the dephosphorylation narrow range of Ca2ϩ. Interestingly, this switch-like response was activity of protein phosphatase 1 (PP1). -

(PDE 1 B 1) Correlates with Brain Regions Having Extensive Dopaminergic Innervation
The Journal of Neuroscience, March 1994, 14(3): 1251-l 261 Expression of a Calmodulin-dependent Phosphodiesterase lsoform (PDE 1 B 1) Correlates with Brain Regions Having Extensive Dopaminergic Innervation Joseph W. Polli and Randall L. Kincaid Section on Immunology, Laboratory of Molecular and Cellular Neurobiology, National Institute on Alcohol Abuse and Alcoholism, Rockville, Maryland 20852 Cyclic nucleotide-dependent protein phosphorylation plays PDE implies an important physiological role for Ca2+-regu- a central role in neuronal signal transduction. Neurotrans- lated attenuation of CAMP-dependent signaling pathways mitter-elicited increases in cAMP/cGMP brought about by following dopaminergic stimulation. activation of adenylyl and guanylyl cyclases are downre- [Key words: CAMP, cyclase, striatum, dopamine, basal gulated by multiple phosphodiesterase (PDE) enzymes. In ganglia, DARPP-321 brain, the calmodulin (CaM)-dependent isozymes are the major degradative activities and represent a unique point of Cyclic nucleotides, acting as “second messengers”or via direct intersection between the cyclic nucleotide- and calcium effects, regulate a diverse array of neuronal functions, from ion (Ca*+)-mediated second messenger systems. Here we de- channel conductance to gene expression. Hydrolysis of 3’,5’- scribe the distribution of the PDEl Bl (63 kDa) CaM-depen- cyclic nucleotidesto 5’-nucleosidemonophosphates is the major dent PDE in mouse brain. An anti-peptide antiserum to this mechanismfor decreasingintracellular cyclic nucleotide levels. isoform immunoprecipitated -3O-40% of cytosolic PDE ac- This reaction is catalyzed by cyclic nucleotide phosphodiester- tivity, whereas antiserum to PDElA2 (61 kDa isoform) re- ase (PDE) enzymes that constitute a large superfamily (Beavo moved 60-70%, demonstrating that these isoforms are the and Reifsynder, 1990). -

Compartment-Specific Phosphorylation of Phosducin In
Faculty Scholarship 2007 Compartment-specific hoP sphorylation of Phosducin in Rods Underlies Adaptation to Various Levels of Illumination Hongman Song Marycharmain Belcastro E. J. Young Maxim Sokolov Follow this and additional works at: https://researchrepository.wvu.edu/faculty_publications Digital Commons Citation Song, Hongman; Belcastro, Marycharmain; Young, E. J.; and Sokolov, Maxim, "Compartment-specific hospP horylation of Phosducin in Rods Underlies Adaptation to Various Levels of Illumination" (2007). Faculty Scholarship. 92. https://researchrepository.wvu.edu/faculty_publications/92 This Article is brought to you for free and open access by The Research Repository @ WVU. It has been accepted for inclusion in Faculty Scholarship by an authorized administrator of The Research Repository @ WVU. For more information, please contact [email protected]. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 282, NO. 32, pp. 23613–23621, August 10, 2007 © 2007 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Compartment-specific Phosphorylation of Phosducin in Rods Underlies Adaptation to Various Levels of Illumination* Received for publication, March 7, 2007, and in revised form, June 11, 2007 Published, JBC Papers in Press, June 14, 2007, DOI 10.1074/jbc.M701974200 Hongman Song, Marycharmain Belcastro, E. J. Young, and Maxim Sokolov1 From the Departments of Ophthalmology and Biochemistry, West Virginia University School of Medicine and West Virginia University Eye Institute, Morgantown, West Virginia 26506 Phosducin is a major phosphoprotein of rod photoreceptors specific complex with the ␥ subunits of visual heterotrimeric that interacts with the G␥ subunits of heterotrimeric G pro- G protein, transducin (4, 5), and other heterotrimeric G pro- teins in its dephosphorylated state. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Predicting Coupling Probabilities of G-Protein Coupled Receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J
Published online 30 May 2019 Nucleic Acids Research, 2019, Vol. 47, Web Server issue W395–W401 doi: 10.1093/nar/gkz392 PRECOG: PREdicting COupling probabilities of G-protein coupled receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J. Silvio Gutkind4, Robert B. Russell1,2,* and Francesco Raimondi1,2,* 1CellNetworks, Bioquant, Heidelberg University, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany, 2Biochemie Zentrum Heidelberg (BZH), Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Miyagi 980-8578, Japan and 4Department of Pharmacology and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA Received February 10, 2019; Revised April 13, 2019; Editorial Decision April 24, 2019; Accepted May 01, 2019 ABSTRACT great use in tinkering with signalling pathways in living sys- tems (5). G-protein coupled receptors (GPCRs) control multi- Ligand binding to GPCRs induces conformational ple physiological states by transducing a multitude changes that lead to binding and activation of G-proteins of extracellular stimuli into the cell via coupling to situated on the inner cell membrane. Most of mammalian intra-cellular heterotrimeric G-proteins. Deciphering GPCRs couple with more than one G-protein giving each which G-proteins couple to each of the hundreds receptor a distinct coupling profile (6) and thus specific of GPCRs present in a typical eukaryotic organism downstream cellular responses. Determining these coupling is therefore critical to understand signalling. Here, profiles is critical to understand GPCR biology and phar- we present PRECOG (precog.russelllab.org): a web- macology. Despite decades of research and hundreds of ob- server for predicting GPCR coupling, which allows served interactions, coupling information is still missing for users to: (i) predict coupling probabilities for GPCRs many receptors and sequence determinants of coupling- specificity are still largely unknown. -

Reporting the Implementation of the Three Rs in European Primate and Mouse Research Papers: Are We Making Progress?
ATLA 38, 495–517, 2010 495 Reporting the Implementation of the Three Rs in European Primate and Mouse Research Papers: Are We Making Progress? Katy Taylor British Union for the Abolition of Vivisection, London, UK Summary — It is now more than 20 years since both Council of Europe Convention ETS123 and EU Directive 86/609?EEC were introduced, to promote the implementation of the Three Rs in animal experi- mentation and to provide guidance on animal housing and care. It might therefore be expected that reports of the implementation of the Three Rs in animal research papers would have increased during this period. In order to test this hypothesis, a literature survey of animal-based research was conducted. A ran- domly-selected sample from 16 high-profile medical journals, of original research papers arising from European institutions that featured experiments which involved either mice or primates, were identified for the years 1986 and 2006 (Total sample = 250 papers). Each paper was scored out of 10 for the incidence of reporting on the implementation of Three Rs-related factors corresponding to Replacement (justification of non-use of non-animal methods), Reduction (statistical analysis of the number of animals needed) and Refinement (housing aspects, i.e. increased cage size, social housing, enrichment of cage environment and food; and procedural aspects, i.e. the use of anaesthesia, analgesia, humane endpoints, and training for procedures with positive reinforcement). There was no significant increase in overall reporting score over time, for either mouse or primate research. By 2006, mouse research papers scored an average of 0 out of a possible 10, and primate research papers scored an average of 1.5. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of