Transcriptome Analysis of Induced Pluripotent Stem Cells from Monozygotic Twins Discordant for Trisomy 21
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

Anti-MRPS17 Monoclonal Antibody, Clone FQS23694 (DCABH-6110) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-MRPS17 monoclonal antibody, clone FQS23694 (DCABH-6110) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Product Overview Rabbit monoclonal to MRPS17 Antigen Description Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that belongs to the ribosomal protein S17P family. The encoded protein is moderately conserved between human mitochondrial and prokaryotic ribosomal proteins. Pseudogenes corresponding to this gene are found on chromosomes 1p, 3p, 6q, 14p, 18q, and Xq. Immunogen Recombinant fragment within Human MRPS17. The exact sequence is proprietary.Database link: Q9Y2R5 Isotype IgG Source/Host Rabbit Species Reactivity Human Clone FQS23694 Purity Tissue culture supernatant Conjugate Unconjugated Applications IHC-P, WB, ICC/IF, IP Positive Control HeLa, HepG2 and U937 cell lysate; Human kidney and liver tissue; HeLa cells Format Liquid Size 100 μl Buffer pH: 7.2; Preservative: 0.01% Sodium azide; Constituents: 49% PBS, 0.05% BSA, 50% Glycerol 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved Preservative 0.01% Sodium Azide Storage Store at +4°C short term (1-2 weeks). -

MPV17L2 Is Required for Ribosome Assembly in Mitochondria Ilaria Dalla Rosa1,†, Romina Durigon1,†, Sarah F
8500–8515 Nucleic Acids Research, 2014, Vol. 42, No. 13 Published online 19 June 2014 doi: 10.1093/nar/gku513 MPV17L2 is required for ribosome assembly in mitochondria Ilaria Dalla Rosa1,†, Romina Durigon1,†, Sarah F. Pearce2,†, Joanna Rorbach2, Elizabeth M.A. Hirst1, Sara Vidoni2, Aurelio Reyes2, Gloria Brea-Calvo2, Michal Minczuk2, Michael W. Woellhaf3, Johannes M. Herrmann3, Martijn A. Huynen4,IanJ.Holt1 and Antonella Spinazzola1,* 1MRC National Institute for Medical Research, Mill Hill, London NW7 1AA, UK, 2MRC Mitochondrial Biology Unit, Wellcome Trust-MRC Building, Hills Road, Cambridge CB2 0XY, UK, 3Cell Biology, University of Kaiserslautern, 67663 Kaiserslautern, Germany and 4Centre for Molecular and Biomolecular Informatics, Radboud University Medical Centre, Geert Grooteplein Zuid 26–28, 6525 GA Nijmegen, Netherlands Received January 15, 2014; Revised May 7, 2014; Accepted May 23, 2014 ABSTRACT INTRODUCTION MPV17 is a mitochondrial protein of unknown func- The mammalian mitochondrial proteome comprises 1500 tion, and mutations in MPV17 are associated with or more gene products. The deoxyribonucleic acid (DNA) mitochondrial deoxyribonucleic acid (DNA) mainte- inside mitochondria DNA (mtDNA) contributes only 13 ∼ nance disorders. Here we investigated its most sim- of these proteins, and they make up 20% of the subunits ilar relative, MPV17L2, which is also annotated as of the oxidative phosphorylation (OXPHOS) system, which produces much of the cells energy. All the other proteins a mitochondrial protein. Mitochondrial fractionation -

Cardiac SARS‐Cov‐2 Infection Is Associated with Distinct Tran‐ Scriptomic Changes Within the Heart
Cardiac SARS‐CoV‐2 infection is associated with distinct tran‐ scriptomic changes within the heart Diana Lindner, PhD*1,2, Hanna Bräuninger, MS*1,2, Bastian Stoffers, MS1,2, Antonia Fitzek, MD3, Kira Meißner3, Ganna Aleshcheva, PhD4, Michaela Schweizer, PhD5, Jessica Weimann, MS1, Björn Rotter, PhD9, Svenja Warnke, BSc1, Carolin Edler, MD3, Fabian Braun, MD8, Kevin Roedl, MD10, Katharina Scher‐ schel, PhD1,12,13, Felicitas Escher, MD4,6,7, Stefan Kluge, MD10, Tobias B. Huber, MD8, Benjamin Ondruschka, MD3, Heinz‐Peter‐Schultheiss, MD4, Paulus Kirchhof, MD1,2,11, Stefan Blankenberg, MD1,2, Klaus Püschel, MD3, Dirk Westermann, MD1,2 1 Department of Cardiology, University Heart and Vascular Center Hamburg, Germany. 2 DZHK (German Center for Cardiovascular Research), partner site Hamburg/Kiel/Lübeck. 3 Institute of Legal Medicine, University Medical Center Hamburg‐Eppendorf, Germany. 4 Institute for Cardiac Diagnostics and Therapy, Berlin, Germany. 5 Department of Electron Microscopy, Center for Molecular Neurobiology, University Medical Center Hamburg‐Eppendorf, Germany. 6 Department of Cardiology, Charité‐Universitaetsmedizin, Berlin, Germany. 7 DZHK (German Centre for Cardiovascular Research), partner site Berlin, Germany. 8 III. Department of Medicine, University Medical Center Hamburg‐Eppendorf, Germany. 9 GenXPro GmbH, Frankfurter Innovationszentrum, Biotechnologie (FIZ), Frankfurt am Main, Germany. 10 Department of Intensive Care Medicine, University Medical Center Hamburg‐Eppendorf, Germany. 11 Institute of Cardiovascular Sciences, -

Inhibition of the MID1 Protein Complex
Matthes et al. Cell Death Discovery (2018) 4:4 DOI 10.1038/s41420-017-0003-8 Cell Death Discovery ARTICLE Open Access Inhibition of the MID1 protein complex: a novel approach targeting APP protein synthesis Frank Matthes1,MoritzM.Hettich1, Judith Schilling1, Diana Flores-Dominguez1, Nelli Blank1, Thomas Wiglenda2, Alexander Buntru2,HannaWolf1, Stephanie Weber1,InaVorberg 1, Alina Dagane2, Gunnar Dittmar2,3,ErichWanker2, Dan Ehninger1 and Sybille Krauss1 Abstract Alzheimer’s disease (AD) is characterized by two neuropathological hallmarks: senile plaques, which are composed of amyloid-β (Aβ) peptides, and neurofibrillary tangles, which are composed of hyperphosphorylated tau protein. Aβ peptides are derived from sequential proteolytic cleavage of the amyloid precursor protein (APP). In this study, we identified a so far unknown mode of regulation of APP protein synthesis involving the MID1 protein complex: MID1 binds to and regulates the translation of APP mRNA. The underlying mode of action of MID1 involves the mTOR pathway. Thus, inhibition of the MID1 complex reduces the APP protein level in cultures of primary neurons. Based on this, we used one compound that we discovered previously to interfere with the MID1 complex, metformin, for in vivo experiments. Indeed, long-term treatment with metformin decreased APP protein expression levels and consequently Aβ in an AD mouse model. Importantly, we have initiated the metformin treatment late in life, at a time-point where mice were in an already progressed state of the disease, and could observe an improved behavioral phenotype. These 1234567890 1234567890 findings together with our previous observation, showing that inhibition of the MID1 complex by metformin also decreases tau phosphorylation, make the MID1 complex a particularly interesting drug target for treating AD. -

Supplementary Dataset S2
mitochondrial translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 MTRF1L 4 MRPL50 MRPS18A MRPS17 2 MRPL20 MRPL52 0 MRPL17 MRPS33 MRPS15 −2 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr ITL original ICr ICa ITL ICa ITL original ICr ITL ICr ICa mitochondrial translational elongation MRPL28 MRPS26 6 MRPS21 PTCD3 MRPS18A 4 MRPS17 MRPL20 2 MRPS15 MRPL45 MRPL52 0 MRPS33 MRPL30 −2 MRPS27 AURKAIP1 MRPS10 MRPL42 MRPL19 MRPL18 MRPL3 MRPS6 MRPL24 MRPS35 MRPL9 MRPS18B MRPL41 MRPS2 MRPL34 MRPS5 MRPL44 MRPS23 MRPS25 MRPL50 MRPL17 GADD45GIP1 ERAL1 MRPL37 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 C12orf65 4 MTRF1L MRPL50 MRPS18A 2 MRPS17 MRPL20 0 MRPL52 MRPL17 MRPS33 −2 MRPS15 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational elongation DIO2 MRPS18B MRPL41 6 MRPS2 MRPL34 GADD45GIP1 4 ERAL1 MRPL37 2 MRPS10 MRPL42 MRPL19 0 MRPL30 MRPS27 AURKAIP1 −2 MRPL18 MRPL3 MRPS6 MRPS35 MRPL9 EEF2K MRPL50 MRPS5 MRPL44 MRPS23 MRPS25 MRPL24 MRPS33 MRPL52 EIF5A2 MRPL17 SECISBP2 MRPS15 MRPL45 MRPS18A MRPS17 MRPL20 MRPL28 MRPS26 MRPS21 PTCD3 ITB ITB ITB ITB ICa ICr ICr ITL original ITL ICa ICa ITL ICr ICr ICa original -

Fhl1p Protein, a Positive Transcription Factor in Pichia Pastoris, Enhances
Zheng et al. Microb Cell Fact (2019) 18:207 https://doi.org/10.1186/s12934-019-1256-0 Microbial Cell Factories RESEARCH Open Access Fhl1p protein, a positive transcription factor in Pichia pastoris, enhances the expression of recombinant proteins Xueyun Zheng1,2, Yimin Zhang1,2, Xinying Zhang1,2, Cheng Li1,2, Xiaoxiao Liu1,2, Ying Lin1,2* and Shuli Liang1,2* Abstract Background: The methylotrophic yeast Pichia pastoris is well-known for the production of a broad spectrum of functional types of heterologous proteins including enzymes, antigens, engineered antibody fragments, and next gen protein scafolds and many transcription factors are utilized to address the burden caused by the high expression of heterologous proteins. In this article, a novel P. pastoris transcription factor currently annotated as Fhl1p, an activator of ribosome biosynthesis processing, was investigated for promoting the expression of the recombinant proteins. Results: The function of Fhl1p of P. pastoris for improving the expression of recombinant proteins was verifed in strains expressing phytase, pectinase and mRFP, showing that the productivity was increased by 20–35%. RNA-Seq was used to study the Fhl1p regulation mechanism in detail, confrming Fhl1p involved in the regulation of rRNA pro- cessing genes, ribosomal small/large subunit biogenesis genes, Golgi vesicle transport genes, etc., which contributed to boosting the expression of foreign proteins. The overexpressed Fhl1p strain exhibited increases in the polysome and monosome levels, showing improved translation activities. Conclusion: This study illustrated that the transcription factor Fhl1p could efectively enhance recombinant protein expression in P. pastoris. Furthermore, we provided the evidence that overexpressed Fhl1p was related to more active translation state. -

Transcriptomic and Proteomic Landscape of Mitochondrial
TOOLS AND RESOURCES Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals Inge Ku¨ hl1,2†*, Maria Miranda1†, Ilian Atanassov3, Irina Kuznetsova4,5, Yvonne Hinze3, Arnaud Mourier6, Aleksandra Filipovska4,5, Nils-Go¨ ran Larsson1,7* 1Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany; 2Department of Cell Biology, Institute of Integrative Biology of the Cell (I2BC) UMR9198, CEA, CNRS, Univ. Paris-Sud, Universite´ Paris-Saclay, Gif- sur-Yvette, France; 3Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 4Harry Perkins Institute of Medical Research, The University of Western Australia, Nedlands, Australia; 5School of Molecular Sciences, The University of Western Australia, Crawley, Australia; 6The Centre National de la Recherche Scientifique, Institut de Biochimie et Ge´ne´tique Cellulaires, Universite´ de Bordeaux, Bordeaux, France; 7Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden Abstract Dysfunction of the oxidative phosphorylation (OXPHOS) system is a major cause of human disease and the cellular consequences are highly complex. Here, we present comparative *For correspondence: analyses of mitochondrial proteomes, cellular transcriptomes and targeted metabolomics of five [email protected] knockout mouse strains deficient in essential factors required for mitochondrial DNA gene (IKu¨ ); expression, leading to OXPHOS dysfunction. Moreover, -

LETM1 Couples Mitochondrial DNA Metabolism and Nutrient Preference
Research Article LETM1 couples mitochondrial DNA metabolism and nutrient preference Romina Durigon1, Alice L Mitchell1,†, Aleck WE Jones1,†, Andreea Manole2,3, Mara Mennuni1, Elizabeth MA Hirst4, Henry Houlden2,3, Giuseppe Maragni5, Serena Lattante5, Paolo Niccolo’ Doronzio5, Ilaria Dalla Rosa1, Marcella Zollino5, Ian J Holt1,6,7 & Antonella Spinazzola1,2,* Abstract transcribed from mitochondrial DNA (mtDNA) and synthesized by the mitochondrial ribosomes (mitoribosomes), via a highly special- The diverse clinical phenotypes of Wolf–Hirschhorn syndrome ized system that is tightly coupled to the inner mitochondrial (WHS) are the result of haploinsufficiency of several genes, one of membrane (IMM; Liu & Spremulli, 2000). In yeasts, the proposed which, LETM1, encodes a protein of the mitochondrial inner machinery of co-translational membrane insertion includes Oxa1, membrane of uncertain function. Here, we show that LETM1 is mba1, and mdm38 (Ott & Herrmann, 2010). Mdm38 is an integral associated with mitochondrial ribosomes, is required for mitochon- inner membrane protein conserved among eukaryotes, and it is drial DNA distribution and expression, and regulates the activity of suggested to play a role in multiple facets of mitochondrial metabo- an ancillary metabolic enzyme, pyruvate dehydrogenase. LETM1 lism, the precise nature of which remains unclear. Under certain deficiency in WHS alters mitochondrial morphology and DNA orga- conditions, it co-fractionates with the mitoribosome, on buoyant nization, as does substituting ketone bodies for glucose in control density gradients (Frazier et al, 2006; Kehrein et al, 2015), and it is cells. While this change in nutrient availability leads to the death proposed to regulate mitochondrial translation (Bauerschmitt et al, of fibroblasts with normal amounts of LETM1, WHS-derived fibrob- 2010). -

MRPS17 Promotes Invasion and Metastasis Through PI3K/AKT Signal
Journal of Cancer 2021, Vol. 12 4849 Ivyspring International Publisher Journal of Cancer 2021; 12(16): 4849-4861. doi: 10.7150/jca.55719 Research Paper MRPS17 promotes invasion and metastasis through PI3K/AKT signal pathway and could be potential prognostic marker for gastric cancer Wenjie Zhou1,2*, Jun Ouyang1,2*, Junqing Li1,2*, Fangjie Liu4, Tailai An1, Lvjia Cheng1,2, Zi Chong Kuo1, Changhua Zhang1 and Yulong He1,2 1. Digestive Disease Center, The Seventh Affiliated Hospital, Sun Yat-sen University, Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. 2. Department of Gastrointestinal Surgery, The First Affiliated Hospital, Sun Yat-sen University, Zhongshan 2nd Road 58, Yuexiu District, Guangzhou 510080, Guangdong, China. 3. Department of Pathology, The Seventh Affiliated Hospital, Sun Yat-sen University, Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. 4. Department of Hematology, The Seventh Affiliated Hospital, Sun Yat-sen University, 58 Zhongshan 2nd Road 58, Yuexiu District, Guangzhou 510080, Guangdong, China. 5. Scientific Research Centre, The Seventh Affiliated Hospital, Sun Yat-sen University, 628 Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. *These authors contributed equally to this work. Corresponding authors: Changhua Zhang, E-mail: [email protected]; Yulong He, E-mail: [email protected]. © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2020.11.10; Accepted: 2021.05.29; Published: 2021.06.11 Abstract In this study, the molecular mechanisms through which Mitochondrial Ribosomal Protein S17 (MRPS17) contributes to gastric cancer (GC) and its prognostic significance in GC have been explored. -
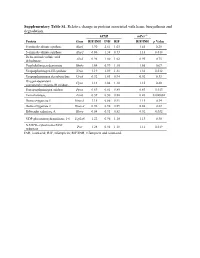
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator-