Foundational and Applied Statistics for Biologists Using R Electronic Appendix (Introduction to R)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mobile Phones and Cloud Computing
Mobile phones and cloud computing A quantitative research paper on mobile phone application offloading by cloud computing utilization Oskar Hamrén Department of informatics Human Computer Interaction Master’s programme Master thesis 2-year level, 30 credits SPM 2012.07 Abstract The development of the mobile phone has been rapid. From being a device mainly used for phone calls and writing text messages the mobile phone of today, or commonly referred to as the smartphone, has become a multi-purpose device. Because of its size and thermal constraints there are certain limitations in areas of battery life and computational capabilities. Some say that cloud computing is just another buzzword, a way to sell already existing technology. Others claim that it has the potential to transform the whole IT-industry. This thesis is covering the intersection of these two fields by investigating if it is possible to increase the speed of mobile phones by offloading computational heavy mobile phone application functions by using cloud computing. A mobile phone application was developed that conducts three computational heavy tests. The tests were run twice, by not using cloud computing offloading and by using it. The time taken to carry out the tests were saved and later compared to see if it is faster to use cloud computing in comparison to not use it. The results showed that it is not beneficial to use cloud computing to carry out these types of tasks; it is faster to use the mobile phone. 1 Table of Contents Abstract ..................................................................................................................................... 1 Table of Contents ..................................................................................................................... 2 1. Introduction .......................................................................................................................... 5 1.1 Previous research ........................................................................................................................ -

Editors Desk ...2
The content of this magazine is released under the Creative Commons Attribution-Share Alike 3.0 Unported license. For more information visit user http://creativecommons.org/licenses/by-sa/3.0 TM Issue #1 - April 2009 EDITORS DESK ................................ 2 COMMUNITY NEWS ........................ 3 CHOOSING A DE/WM ...................... 4 HARDENING SSH IN 60 SECONDS .................................... 6 GAMERS CORNER .......................... 9 TIPS & TRICKS ............................... 10 PIMP MY ARCH .............................. 11 SOFTWARE REVIEW ......................12 Q&A ..................................................14 EEDDIITTOORRSS DDEESSKK Welcome to the first issue of Arch User Magazine! ARCH USER STAFF Daniel Griffiths (Ghost1227) ........... Editor ello, and thank you for picking up issue #1 of Arch User Magazine! While David Crouse (Crouse) .......... Contributor the vast majority of you probably know me (or have at least seen me H around the forums), I feel that I should take a moment to introduce myself. My name is Daniel Griffiths, and I am a 26-year-old independent contractor in Delaware, US. Throughout my life, I have wandered through various UNIX/Linux systems including (but not limited to) MINIX, RedHat, Mandrake, Slackware, Gentoo, Debian, and even two home made distributions based on Linux From Scratch. I finally found Arch in 2007 and instantly fell in love with its elegant simplicity. Some of our more attentive readers may note that Arch already has a monthly newsletter. With the existence of the aformentioned newsletter, what is the point of adding another news medium to the mix? Fear not, newsletter readers, I have no intention of letting Arch User Magazine take the place of the newsletter. In fact, Arch User Magazine and the newsletter are intended to fill two very different needs in the Arch community. -

Download (4MB)
Jurnal SIMETRIS, Vol 6 No 2 November 2015 ISSN: 2252-4983 REMASTERING LIVE USB UNTUK ”LAMP” PADA FAKULTAS SAINS DAN TEKNOLOGI PALEMBANG Klaudius Jevanda B.S. Fakultas Sains dan Teknologi, Program Studi Informatika Universitas Katolik Musi Charitas Email: [email protected] ABSTRAK Penelitian ini bertujuan untuk membuat distribusi Linux bernama Lubuntu yang memfokuskan diri pada desktop yang ringan serta ditujukan untuk menjadi lingkungan web server yang free dalam bentuk live USB. Dimana, penelitian ini menjelaskan tentang desain dan implementasi dari distribusi Linux Lubuntu itu sendiri yang nantinya bisa terus diperbaiki, disempurnakan dan dimungkinkan untuk dimodifikasi serta dipelajari oleh pihak lain. Lubuntu dikembangkan dengan memodifikasi dari Linux Lubuntu 14.04 dari tahap penambahan program, penghapusan program dan konfigurasi sampai pada tahap pembuatan Live USB untuk LAMP (Linux Apache Mysql PHP) menggunakan metode remastering. Hasil dari penelitian ini berupa Live USB yang berisi tool untuk lingkungan web server. Tool utama dalam Live USB diantaranya adalah phpmyadmin, gimp, inkscape, dan bluefish. Keluaran penelitian ini, diharapkan bisa digunakan sebagai sistem operasi dan dikhususkan dalam lingkungan web server yang nyaman untuk dipergunakan dalam proses belajar mengajar pada matakuliah sistem operasi, pemrograman basis web I, dan pemrograman basis web II di Fakultas Sains dan Teknologi Universitas Katolik Musi Charitas palembang. Kata kunci: linux, web server, LAMP, remastering, live usb. ABSTRACT The research aims to make the linux distribution namely Lubuntu that focuses on lightweight desktop environment and to create a free Live USB environment. This research explains the design and implementation the Linux Lubuntu distribution which will be continuously improved, modified, and learned by other. -

LIFE Packages
LIFE packages Index Office automation Desktop Internet Server Web developpement Tele centers Emulation Health centers Graphics High Schools Utilities Teachers Multimedia Tertiary schools Programming Database Games Documentation Internet - Firefox - Browser - Epiphany - Nautilus - Ftp client - gFTP - Evolution - Mail client - Thunderbird - Internet messaging - Gaim - Gaim - IRC - XChat - Gaim - VoIP - Skype - Videomeeting - Gnome meeting - GnomeBittorent - P2P - aMule - Firefox - Download manager - d4x - Telnet - Telnet Web developpement - Quanta - Bluefish - HTML editor - Nvu - Any text editor - HTML galerie - Album - Web server - XAMPP - Collaborative publishing system - Spip Desktop - Gnome - Desktop - Kde - Xfce Graphics - Advanced image editor - The Gimp - KolourPaint - Simple image editor - gPaint - TuxPaint - CinePaint - Video editor - Kino - OpenOffice Draw - Vector vraphics editor - Inkscape - Dia - Diagram editor - Kivio - Electrical CAD - Electric - 3D modeller/render - Blender - CAD system - QCad Utilities - Calculator - gCalcTool - gEdit - gxEdit - Text editor - eMacs21 - Leafpad - Application finder - Xfce4-appfinder - Desktop search tool - Beagle - File explorer - Nautilus -Archive manager - File-Roller - Nautilus CD Burner - CD burner - K3B - GnomeBaker - Synaptic - System updates - apt-get - IPtables - Firewall - FireStarter - BackupPC - Backup - Amanda - gnome-terminal - Terminal - xTerm - xTerminal - Scanner - Xsane - Partition editor - gParted - Making image of disks - Partitimage - Mirroring over network - UDP Cast -

ODROID Magazine, Published Monthly at Is Your Source for All Things Odroidian
Weather Board Application • Open Media Vault • Installing Node.js Year One Issue #11 Nov 2014 ODROIDMagazine 3 X U VIRTUALIZE NOW! DISCOVER A UNIVERSE OF POSSIBILITIES WITH KVM TECHNOLOGY BOINC THE DISTRIBUTED PROCESSING PLATFORM THAT MAKES THE MOST OF AN ODROID’S LOW POWER CONSUMPTION • OS SPOTLIGHT: CODE MONKEY BOINC MONSTER: • LINUX GAMING: DOSBOX A WHOPPING • UNMANNED GROUND VEHICLE: 135W CLUSTER GPS NAVIGATION PROGRAMMING WITH 96 CORES What we stand for. We strive to symbolize the edge of technology, future, youth, humanity, and engineering. Our philosophy is based on Developers. And our efforts to keep close relationships with developers around the world. For that, you can always count on having the quality and sophistication that is the hallmark of our products. Simple, modern and distinctive. So you can have the best to accomplish everything you can dream of. We are now shipping the ODROID U3 devices to EU countries! Come and visit our online store to shop! Address: Max-Pollin-Straße 1 85104 Pförring Germany Telephone & Fax phone : +49 (0) 8403 / 920-920 email : [email protected] Our ODROID products can be found at http://bit.ly/1tXPXwe EDITORIAL The exciting news this month is that ODROIDs are now available for sale in the United States from http://www.ameridroid.com! Based in California, Ameridroid offers affordable shipping for domestic cus- tomers, and US residents will receive packages much faster. Here is an excerpt from their website: “Maybe your story is the same. Santa answered a 7-year-old’s letter with a shiny soldering iron. Before long, he was taking apart electronics and salvaging the parts to make a crystal radio. -

Awoken Icon Theme - Installation & Customizing Instructions 1
Awoken Icon Theme - Installation & Customizing Instructions 1 AWOKEN ICON THEME Installation & Customizing Instructions Alessandro Roncone mail: [email protected] homepage: http://alecive.deviantart.com/ Awoken homepage (GNOME Version): link kAwoken homepage (KDE Version): link Contents 1 Iconset Credits 3 2 Copyright 3 3 Installation 3 3.1 GNOME........................................................3 3.2 KDE..........................................................4 4 Customizing Instructions 4 4.1 GNOME........................................................4 4.2 KDE..........................................................5 5 Overview of the customization script6 5.1 How to customize a single iconset..........................................7 6 Customization options 8 6.1 Folder types......................................................8 6.2 Color-NoColor.................................................... 11 6.3 Distributor Logos................................................... 11 6.4 Trash types...................................................... 11 6.5 Other Options.................................................... 11 6.5.1 Gedit icon................................................... 11 6.5.2 Computer icon................................................ 11 6.5.3 Home icon................................................... 11 6.6 Deprecated...................................................... 12 7 How to colorize the iconset 13 8 Icons that don't want to change (but I've drawed) 14 9 Conclusions 15 9.1 Changelog...................................................... -
Manuale Nel Codice Sarà Necessario
Della serie Les tutoriels libres presentati dal sito FRAMASOFT Framasoft « Partire da Windows per scoprire il software libero... » http://www.framasoft.net Tutorial Framasoft N|VUN|VU Ze|N tutorial Programma: Nvu Autore(i): Lindows Inc. Piattaforma(e): Linux, Windows, Mac Versione: 0.41 it Licenza: mpl/gpl/lgpl Sito italiano: http://www.sanavia.it/nvuitalia Sito inglese: http://nvu.com di Fun Sun (jean-marc juin1) 14/05/04 (prima versione) 24/06/04 (correzioni + aggiornamento 0.3) 25/08/04-->19/10/04 (miglioramenti generali ) Correttori Christian (Choul) Goofy Versione francese 0.41 27.10.04 Traduzione italiana 0.3 di: Giulio Bellezza ([email protected]) 15.11.04 Traduzione, revisione e aggiornamento 0.41 Andrea Sanavia 15.11.04 Correzione e verifica: Riccardo Fusaroli Versione italiana 0.41 beta 1 Pubblicato con licenza Creative Commons By-NonCommercial-ShareAlike http: //creativecommons.org/licenses/b y-nc-sa/1 .0/ Questo documento sarà visualizzato meglio con il carattere bitstream, prelevabile a questo indirizzo: http://ftp.gnome.org/pub/GNOME/sources/ttf-bitstream-vera/1.10/ 1) Patronimo fisico in omaggio a questo mondo rassegnato in ricerca di tempo e di leggi. Un giorno, forse, tu mi autorizzerai a chiudere il suono di queste parole in una chiesa affinché tu prenda coscienza dell'inesistenza dell'alfabeto... Http://www.framasoft.net 2/182 Tutorial Framasoft SOMMARIO Tabella degli argomenti A ) Introduzione ................................................................................................ 6 1) Creare pagine Web e siti -

Technical Notes All Changes in Fedora 13
Fedora 13 Technical Notes All changes in Fedora 13 Edited by The Fedora Docs Team Copyright © 2010 Red Hat, Inc. and others. The text of and illustrations in this document are licensed by Red Hat under a Creative Commons Attribution–Share Alike 3.0 Unported license ("CC-BY-SA"). An explanation of CC-BY-SA is available at http://creativecommons.org/licenses/by-sa/3.0/. The original authors of this document, and Red Hat, designate the Fedora Project as the "Attribution Party" for purposes of CC-BY-SA. In accordance with CC-BY-SA, if you distribute this document or an adaptation of it, you must provide the URL for the original version. Red Hat, as the licensor of this document, waives the right to enforce, and agrees not to assert, Section 4d of CC-BY-SA to the fullest extent permitted by applicable law. Red Hat, Red Hat Enterprise Linux, the Shadowman logo, JBoss, MetaMatrix, Fedora, the Infinity Logo, and RHCE are trademarks of Red Hat, Inc., registered in the United States and other countries. For guidelines on the permitted uses of the Fedora trademarks, refer to https:// fedoraproject.org/wiki/Legal:Trademark_guidelines. Linux® is the registered trademark of Linus Torvalds in the United States and other countries. Java® is a registered trademark of Oracle and/or its affiliates. XFS® is a trademark of Silicon Graphics International Corp. or its subsidiaries in the United States and/or other countries. All other trademarks are the property of their respective owners. Abstract This document lists all changed packages between Fedora 12 and Fedora 13. -

1. Why POCS.Key
Symptoms of Complexity Prof. George Candea School of Computer & Communication Sciences Building Bridges A RTlClES A COMPUTER SCIENCE PERSPECTIVE OF BRIDGE DESIGN What kinds of lessonsdoes a classical engineering discipline like bridge design have for an emerging engineering discipline like computer systems Observation design?Case-study editors Alfred Spector and David Gifford consider the • insight and experienceof bridge designer Gerard Fox to find out how strong the parallels are. • bridges are normally on-time, on-budget, and don’t fall ALFRED SPECTORand DAVID GIFFORD • software projects rarely ship on-time, are often over- AS Gerry, let’s begin with an overview of THE DESIGN PROCESS bridges. AS What is the procedure for designing and con- GF In the United States, most highway bridges are budget, and rarely work exactly as specified structing a bridge? mandated by a government agency. The great major- GF It breaks down into three phases: the prelimi- ity are small bridges (with spans of less than 150 nay design phase, the main design phase, and the feet) and are part of the public highway system. construction phase. For larger bridges, several alter- There are fewer large bridges, having spans of 600 native designs are usually considered during the Blueprints for bridges must be approved... feet or more, that carry roads over bodies of water, preliminary design phase, whereas simple calcula- • gorges, or other large obstacles. There are also a tions or experience usually suffices in determining small number of superlarge bridges with spans ap- the appropriate design for small bridges. There are a proaching a mile, like the Verrazzano Narrows lot more factors to take into account with a large Bridge in New Yor:k. -

Manual De Instalación De Kompozer
UNIVERSIDAD NACIONAL DE CHIMBORAZO FACULTAD DE CIENCIAS DE LA EDUCACIÓN HUMANAS Y TECNOLOGÍAS TÍTULO DE TESIS: “ANÁLISIS, DISEÑO E IMPLEMENTACIÓN DE UN SITIO WEB PARA LA ESCUELA DE INFORMÁTICA APLICADA A LA EDUCACIÓN DE LA UNIVERSIDAD NACIONAL DE CHIMBORAZO UTILIZANDO SOFTWARE LIBRE” Trabajo presentado como requisito para obtener el título de Licenciado en Ciencias de la Educación especialidad Informática Aplicada a la Educación. Autores: Reinoso Quishpi Benjamín Rodrigo benjamí[email protected] Cepeda Zambrano Wilson Leonardo [email protected] Director de Tesis: Ing. Jorge Fernández Acevedo Riobamba – Ecuador 2014 CERTIFICACIÓN Certifico que el presente trabajo fue realizado en su totalidad por los Señores. Wilson Leonardo Cepeda Zambrano y Benjamín Rodrigo Reinoso Quishpi, como requerimiento parcial a la obtención del título de LICENCIADO EN CIENCIAS DE LA EDUCACIÓN, ESPECIALIDAD INFORMÁTICA APLICADA LA EDUCACIÓN. Riobamba,………………………………… Ing. Jorge Fernández Acevedo DIRECTOR DEL PROYECTO DE TESIS UNIVERSIDAD NACIONAL DE CHIMBORAZO FACULTAD DE CIENCIAS DE LA EDUCACIÓN HUMANAS Y TECNOLOGÍAS ESCUELA DE INFORMÁTICA APLICADA A LA EDUCACIÓN TÍTULO DE TESIS: “ANÁLISIS, DISEÑO E IMPLEMENTACIÓN DE UN SITIO WEB PARA LA ESCUELA DE INFORMÁTICA APLICADA A LA EDUCACIÓN DE LA UNIVERSIDAD NACIONAL DE CHIMBORAZO UTILIZANDO SOFTWARE LIBRE” Trabajo de Licenciatura en la especialidad de “Informática Aplicada a la Educación” Autores: Reinoso Quishpi Benjamín Rodrigo Cepeda Zambrano Wilson Leonardo Aprobado en nombre de la Universidad Nacional de Chimborazo por el siguiente jurado examinador a los……..días del mes de……………………del año 2014. Ing. María Eugenia Solís M. …………………………….. PRESIDENTA DEL TRIBUNAL FIRMA Ms. Patricio Tobar. ……………………………... MIEMBRO DEL TRIBUNAL FIRMA Ing. Jorge Fernández A. ……………………………… TUTOR DE TESIS FIRMA NOTA: ………. -
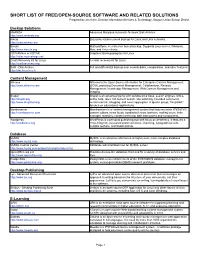
SHORT LIST of FREE/OPEN-SOURCE SOFTWARE and RELATED SOLUTIONS Prepared by Jim Klein, Director Information Services & Technology, Saugus Union School District
SHORT LIST OF FREE/OPEN-SOURCE SOFTWARE AND RELATED SOLUTIONS Prepared by Jim Klein, Director Information Services & Technology, Saugus Union School District Backup Solutions AMANDA Advanced Maryland Automatic Network Disk Archiver http://www.amanda.org Arkeia Enterprise-class network backup for Linux and Unix networks. http://www.arkeia.com Bacula Multi-platform, client/server based backup. Supports Linux server, Windows, http://www.bacula.org Mac, and Linux clients. CDTARchive or CDTAR Graphical Backup program for Linux http://cdtar.sourceforge.net Crash Recovery Kit for Linux A crash recovery kit for Linux http://crashrecovery.org DAR - Disk Archive Full and differential backup over several disks, compression, and other features http://dar.linux.free.fr Content Management Alfresco Alfresco is the Open Source Alternative for Enterprise Content Management http://www.alfresco.com (ECM), providing Document Management, Collaboration, Records Management, Knowledge Management, Web Content Management and Imaging. Drupal Drupal is an advanced portal with collaborative book, search engines, URLs, http://drupal.org online help, roles, full content search, site watching, threaded comments, http://www.drupaled.org version control, blogging, and news aggregator. A special group, “Drupaled,” focuses on educational applications. Mamboserver Mamboserver is a content management system that features inline WYSIWYG http://mamboserver.com content editors, news feeds, syndicated news, banners, mailing users, links manager, statistics, content archiving, date bamodules and components. Wordpress WordPress is a personal publishing tool with focus on aesthetics. It features a http://wordpress.org cross-blog tool, password protected posts, importing, typographical aids, multiple authors, and bookmarklets. Database MySQL MySQL is an attractive alternative to higher-cost, more complex database http://www.mysql.com technology. -

Linux Software Catalog 2018
Linux Software Catalog 2018 This is a list of software I use on a daily bases and consider the best of the best. Although most can be found in the software tool provided by your distro, some may be outdated or froze for stability. I tend to use Flatpak to get somethings, like Krita, so I am using the latest versions. Flatpak apps are installed locally and do not interfere with the underlying system or package manager. I also go directly to the source for others. If you can’t find an app listed here in the package manager or Flatpak Google the name. You can get flatpak apps here: https://flathub.org/home I use Budgie on top of Pop! OS. I use Pop! OS because I am on a System76 laptop and drivers are excellent. I do like Budgie over Gnome, Cinnamon, Mate, etc. regardless of underlying Linux flavor as it’s closer to OS X than the others. I like the work-flow. My current desktop: To get a bigger selection of software open the terminal and type: sudo apt install synaptic Provide the password you set at install. Do not lose that password as it is the only way to manage everything on the system. GRadio (I can't live without this app.) GRadio is installed using Flatpak. It is not available in the Ubuntu repos. https://flathub.org/apps/details/de.haeckerfelix.gradio Open the terminal and type: flatpak install flathub de.haeckerfelix.gradio Then type (should show up in the menu, maybe logout and in if not): flatpak run de.haeckerfelix.gradio Rhythmbox can also play internet radio, gradio just does it better.