SUPPLEMENT Identification of Genes Associated with Chemotherapy Cross-Resistance And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nuclear and Mitochondrial Genome Defects in Autisms
UC Irvine UC Irvine Previously Published Works Title Nuclear and mitochondrial genome defects in autisms. Permalink https://escholarship.org/uc/item/8vq3278q Journal Annals of the New York Academy of Sciences, 1151(1) ISSN 0077-8923 Authors Smith, Moyra Spence, M Anne Flodman, Pamela Publication Date 2009 DOI 10.1111/j.1749-6632.2008.03571.x License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California THE YEAR IN HUMAN AND MEDICAL GENETICS 2009 Nuclear and Mitochondrial Genome Defects in Autisms Moyra Smith, M. Anne Spence, and Pamela Flodman Department of Pediatrics, University of California, Irvine, California In this review we will evaluate evidence that altered gene dosage and structure im- pacts neurodevelopment and neural connectivity through deleterious effects on synap- tic structure and function, and evidence that the latter are key contributors to the risk for autism. We will review information on alterations of structure of mitochondrial DNA and abnormal mitochondrial function in autism and indications that interactions of the nuclear and mitochondrial genomes may play a role in autism pathogenesis. In a final section we will present data derived using Affymetrixtm SNP 6.0 microar- ray analysis of DNA of a number of subjects and parents recruited to our autism spectrum disorders project. We include data on two sets of monozygotic twins. Col- lectively these data provide additional evidence of nuclear and mitochondrial genome imbalance in autism and evidence of specific candidate genes in autism. We present data on dosage changes in genes that map on the X chromosomes and the Y chro- mosome. -

Genetics of Familial Non-Medullary Thyroid Carcinoma (FNMTC)
cancers Review Genetics of Familial Non-Medullary Thyroid Carcinoma (FNMTC) Chiara Diquigiovanni * and Elena Bonora Unit of Medical Genetics, Department of Medical and Surgical Sciences, University of Bologna, 40138 Bologna, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-051-208-8418 Simple Summary: Non-medullary thyroid carcinoma (NMTC) originates from thyroid follicular epithelial cells and is considered familial when occurs in two or more first-degree relatives of the patient, in the absence of predisposing environmental factors. Familial NMTC (FNMTC) cases show a high genetic heterogeneity, thus impairing the identification of pivotal molecular changes. In the past years, linkage-based approaches identified several susceptibility loci and variants associated with NMTC risk, however only few genes have been identified. The advent of next-generation sequencing technologies has improved the discovery of new predisposing genes. In this review we report the most significant genes where variants predispose to FNMTC, with the perspective that the integration of these new molecular findings in the clinical data of patients might allow an early detection and tailored therapy of the disease, optimizing patient management. Abstract: Non-medullary thyroid carcinoma (NMTC) is the most frequent endocrine tumor and originates from the follicular epithelial cells of the thyroid. Familial NMTC (FNMTC) has been defined in pedigrees where two or more first-degree relatives of the patient present the disease in absence of other predisposing environmental factors. Compared to sporadic cases, FNMTCs are often multifocal, recurring more frequently and showing an early age at onset with a worse outcome. FNMTC cases Citation: Diquigiovanni, C.; Bonora, E. -

Genetic Influences on Brain Gene Expression in Rats Selected for Tameness and Aggression
Genetic Influences on Brain Gene Expression in Rats Selected for Tameness and Aggression Henrike O. Heyne*,§, Susann Lautenschläger§, Ronald Nelson†, François Besnier†,‡, Maxime Rotival**, Alexander Cagan*, Rimma Kozhemyakina§§, Irina Z. Plyusnina§§, Lyudmila Trut§§, Örjan Carlborg†, Enrico Petretto**, Leonid Kruglyak††,‡‡,***, Svante Pääbo*, Torsten Schöneberg§, Frank W. Albert*,†† * Department of Evolutionary Genetics, Max Planck Institute for Evolutionary Anthropology, Deutscher Platz 6, 04103 Leipzig, Germany § Institute for Biochemistry, University of Leipzig, Medical Faculty, Johannisallee 30, 04103 Leipzig † Swedish University of Agricultural Sciences, Department of Clinical Sciences, Division of Computational Genetics, Mailing address: Box 7078 SE-75007 Uppsala Sweden ‡ Section of Population Genetics and Ecology, Institute of Marine Research, Bergen, Norway, Havforskningsinstituttet, Postboks 1870, Nordnes 5817, Bergen Norway ** MRC Clinical Sciences Centre, Faculty of Medicine, Imperial College London, Du Cane Road, London W12 0NN, UK §§ Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia †† Department of Human Genetics, University of California, Los Angeles, Gonda Center, 695 Charles E. Young Drive South, Los Angeles, CA 90095, USA ‡‡ Department of Biological Chemistry, University of California, Los Angeles, Gonda Center, 695 Charles E. Young Drive South, Los Angeles, CA 90095, USA *** Howard Hughes Medical Institute, University of California, Los Angeles, Gonda Center, 695 Charles E. Young Drive South, Los Angeles, CA 90095, USA deceased: I.Z.P. Irina Z. Plyusnina . 1 ABSTRACT Inter-individual differences in many behaviors are partly due to genetic differences, but the identification of the genes and variants that influence behavior remains challenging. Here, we studied an F2 intercross of two outbred lines of rats selected for tame and aggressive behavior towards humans for more than 64 generations. -

The Putative Tumor Suppressor Gene GLTSCR2 Induces PTEN-Modulated Cell Death
Cell Death and Differentiation (2007) 14, 1872–1879 & 2007 Nature Publishing Group All rights reserved 1350-9047/07 $30.00 www.nature.com/cdd The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death J-H Yim1,2, Y-J Kim1,2, J-H Ko1,2, Y-E Cho1,2, S-M Kim1,2, J-Y Kim1,2, S Lee1,2 and J-H Park*,1,2 Glioma tumor suppressor candidate region gene 2 (GLTSCR2/PICT-1) is localized within the well-known 1.4-Mb tumor suppressive region of chromosome 19q, which is frequently altered in various human tumors, including diffuse gliomas. Aside from its localization on the chromosome, several lines of evidence, such as PTEN phosphorylation, support that GLTSCR2 partakes in the suppression of tumor growth and development. However, much remains unknown about the molecular mechanisms of the tumor suppressive activity of GLTSCR2. The purpose of this study was to investigate the molecular mechanisms of GLTSCR2 in cell death pathways in association with its binding partner PTEN. In this work, we show that GLTSCR2 is a nucleus-localized protein with a discrete globular expression pattern. In addition to phosphorylating PTEN, GLTSCR2 induces caspase-independent PTEN-modulated apoptotic cell death when overexpressed. However, the cytotoxic activity of GLTSCR2 is independent of its ability to phosphorylate PTEN, suggesting that the GLTSCR2-induced cell death pathway is divergent from PTEN-induced death pathways. Our results suggest that the induction of PTEN-modulated apoptosis is one of the putative mechanisms of tumor suppressive activity -

Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signifcance and Impact on Clinical Features
Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signicance and Impact on Clinical Features Fadwa Said Abdelazim Mohamed ( [email protected] ) Cairo University Roxan E Shak National Cancer Institute, Cairo University Naglaa M. Hassan National Cancer Institute, Cairo University Research article Keywords: Acute myeloid leukemia, RUNX1, mutation, gene expression, Egypt Posted Date: April 7th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-20926/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/20 Abstract Background Acute myeloid leukemia represents the highest percentage of all adult acute leukemia variants. Runt-related transcription factor1 (RUNX1), a transcription factor with a known tumor suppressor function was recently reported as a tumor promotor in AML. We investigated the role of RUNX1 geneexpression levelin Egyptian AML patients and delineateits clinical signicance. Methods This study recruited 91 AML patients that were recently diagnosed at our hospital with 14 healthy age- and sex-matched donors of bone marrow transplantation unit. We measured RUNX1 gene expression level using reverse transcription–quantitative polymerase chain reaction. Results We found that RUNX1 gene expression level was signicantly higher compared to the control group (p < 0.001). Patients with FLT3 mutations had the higher expression level of RUNX1 (p=0.023). The male patients expressed signicantly higher level of RUNX1 (p=0.046). Conclusion The RUNX1 gene expression may serve as a diagnostic marker in Egyptian AML patients. Its relation to FLT3 may give clue that patients carrying this mutation may benet fromnew treatments that target RUNX1 in the future. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -
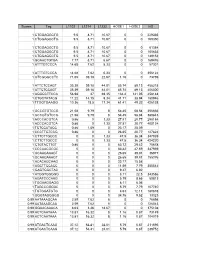
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

(12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer Et Al
USOO8440393B2 (12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer et al. (45) Date of Patent: May 14, 2013 (54) PRO-ANGIOGENIC GENES IN OVARIAN OTHER PUBLICATIONS TUMORENDOTHELIAL CELL, SOLATES Boyd (The Basic Science of Oncology, 1992, McGraw-Hill, Inc., p. (75) Inventors: Michael J. Birrer, Mt. Airy, MD (US); 379). Tomas A. Bonome, Washington, DC Tockman et al. (Cancer Res., 1992, 52:2711s-2718s).* (US); Anil Sood, Pearland, TX (US); Pritzker (Clinical Chemistry, 2002, 48: 1147-1150).* Chunhua Lu, Missouri City, TX (US) Benedict et al. (J. Exp. Medicine, 2001, 193(1) 89-99).* Jiang et al. (J. Biol. Chem., 2003, 278(7) 4763-4769).* (73) Assignees: The United States of America as Matsushita et al. (FEBS Letters, 1999, vol. 443, pp. 348-352).* Represented by the Secretary of the Singh et al. (Glycobiology, 2001, vol. 11, pp. 587-592).* Department of Health and Human Abbosh et al. (Cancer Res. Jun. 1, 2006 66:5582-55.91 and Supple Services, Washington, DC (US); The mental Figs. S1-S7).* University of MD Anderson Cancer Zhai et al. (Chinese General Practice Aug. 2008, 11(8A): 1366 Center, Houston, TX (US) 1367).* Lu et al. (Cancer Res. Feb. 15, 2007, 64(4): 1757-1768).* (*) Notice: Subject to any disclaimer, the term of this Bagnato et al., “Activation of Mitogenic Signaling by Endothelin 1 in patent is extended or adjusted under 35 Ovarian Carcinoma Cells', Cancer Research, vol. 57, pp. 1306-1311, U.S.C. 154(b) by 194 days. 1997. Bouras et al., “Stanniocalcin 2 is an Estrogen-responsive Gene (21) Appl. -

Download 20190410); Fragmentation for 20 S
ARTICLE https://doi.org/10.1038/s41467-020-17387-y OPEN Multi-layered proteomic analyses decode compositional and functional effects of cancer mutations on kinase complexes ✉ Martin Mehnert 1 , Rodolfo Ciuffa1, Fabian Frommelt 1, Federico Uliana1, Audrey van Drogen1, ✉ ✉ Kilian Ruminski1,3, Matthias Gstaiger1 & Ruedi Aebersold 1,2 fi 1234567890():,; Rapidly increasing availability of genomic data and ensuing identi cation of disease asso- ciated mutations allows for an unbiased insight into genetic drivers of disease development. However, determination of molecular mechanisms by which individual genomic changes affect biochemical processes remains a major challenge. Here, we develop a multilayered proteomic workflow to explore how genetic lesions modulate the proteome and are trans- lated into molecular phenotypes. Using this workflow we determine how expression of a panel of disease-associated mutations in the Dyrk2 protein kinase alter the composition, topology and activity of this kinase complex as well as the phosphoproteomic state of the cell. The data show that altered protein-protein interactions caused by the mutations are asso- ciated with topological changes and affected phosphorylation of known cancer driver pro- teins, thus linking Dyrk2 mutations with cancer-related biochemical processes. Overall, we discover multiple mutation-specific functionally relevant changes, thus highlighting the extensive plasticity of molecular responses to genetic lesions. 1 Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, -

An Optimized Streptavidin-Binding RNA Aptamer for Purification Of
Published online 23 October 2013 Nucleic Acids Research, 2014, Vol. 42, No. 2 e13 doi:10.1093/nar/gkt956 An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins Kathrin Leppek1,2,3 and Georg Stoecklin1,2,3,* 1Helmholtz Junior Research Group Posttranscriptional Control of Gene Expression, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany, 2Zentrum fu¨ r Molekulare Biologie der Universita¨ t Heidelberg (ZMBH), Im Neuenheimer Feld 282, 69120 Heidelberg, Germany and 3DKFZ-ZMBH Downloaded from https://academic.oup.com/nar/article-abstract/42/2/e13/1030103 by guest on 11 March 2019 Alliance Received June 19, 2013; Revised September 6, 2013; Accepted September 27, 2013 ABSTRACT degradation (1,2). The fate of any given mRNA is essen- tially determined by the messenger ribonucleoprotein Determining the composition of messenger (mRNP) complex, i.e. the ensemble of proteins and regu- ribonucleoprotein (mRNP) particles is essential for latory RNAs an mRNA interacts with. Although decades a comprehensive understanding of the complex of research have uncovered specific functions of numerous mechanisms underlying mRNA regulation, but is RNA-binding proteins (RNA-BPs), surprisingly little is technically challenging. Here we present an RNA- known about the composition, heterogeneity and based method to identify RNP components using a dynamics of whole mRNPs. modified streptavidin (SA)-binding RNA aptamer There are essentially two strategies to explore the termed S1m. By optimizing the RNA aptamer S1 in content of mRNPs. On one hand, protein-based structure and repeat conformation, we improved its approaches rely on the purification of a particular RNA- affinity for SA and found a 4-fold repeat of S1m BP by immunoprecipitation together with its associated mRNAs, whose identity can then be determined by (4ÂS1m) to be more efficient than the established cDNA cloning, microarray analysis (RIP-Chip) or deep MS2 and PP7 systems from bacteriophages. -

Candidate Genes, Pathways and Mechanisms for Bipolar (Manic–Depressive) and Related Disorders: an Expanded Convergent Function
Molecular Psychiatry (2004), 1007–1029 & 2004 Nature Publishing Group All rights reserved 1359-4184/04 $30.00 www.nature.com/mp ORIGINAL RESEARCH ARTICLE Candidate genes, pathways and mechanisms for bipolar (manic–depressive) and related disorders: an expanded convergent functional genomics approach CA Ogden1,2,3, ME Rich1,2,3, NJ Schork2, MP Paulus2,3, MA Geyer2,3, JB Lohr2,3, R Kuczenski2,3 and AB Niculescu1,2,3 1Laboratory of Neurophenomics, University of California, San Diego, CA, USA; 2Department of Psychiatry, University of California, San Diego, CA, USA; 3VA San Diego Healthcare System VISN-22 MIRECC, San Diego, CA, USA Identifying genes for bipolar mood disorders through classic genetics has proven difficult. Here, we present a comprehensive convergent approach that translationally integrates brain gene expression data from a relevant pharmacogenomic mouse model (involving treatments with a stimulant—methamphetamine, and a mood stabilizer—valproate), with human data (linkage loci from human genetic studies, changes in postmortem brains from patients), as a bayesian strategy of crossvalidating findings. Topping the list of candidate genes, we have DARPP-32 (dopamine- and cAMP-regulated phosphoprotein of 32 kDa) located at 17q12, PENK (preproenkephalin) located at 8q12.1, and TAC1 (tachykinin 1, substance P) located at 7q21.3. These data suggest that more primitive molecular mechanisms involved in pleasure and pain may have been recruited by evolution to play a role in higher mental functions such as mood. The analysis also revealed other high-probability candidates genes (neurogenesis, neuro- trophic, neurotransmitter, signal transduction, circadian, synaptic, and myelin related), pathways and mechanisms of likely importance in pathophysiology.