Candidate Genes, Pathways and Mechanisms for Bipolar (Manic–Depressive) and Related Disorders: an Expanded Convergent Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Changes in Poly(A) Tail Length Dynamics from the Loss of the Circadian Deadenylase Nocturnin
www.nature.com/scientificreports OPEN Changes in poly(A) tail length dynamics from the loss of the circadian deadenylase Nocturnin Received: 16 February 2015 1,2 2 1 1 1 Accepted: 21 October 2015 Shihoko Kojima , Kerry L. Gendreau , Elaine L. Sher-Chen , Peng Gao & Carla B. Green Published: 20 November 2015 mRNA poly(A) tails are important for mRNA stability and translation, and enzymes that regulate the poly(A) tail length significantly impact protein profiles. There are eleven putative deadenylases in mammals, and it is thought that each targets specific transcripts, although this has not been clearly demonstrated. Nocturnin (NOC) is a unique deadenylase with robustly rhythmic expression and loss of Noc in mice (Noc KO) results in resistance to diet-induced obesity. In an attempt to identify target transcripts of NOC, we performed “poly(A)denylome” analysis, a method that measures poly(A) tail length of transcripts in a global manner, and identified 213 transcripts that have extended poly(A) tails in Noc KO liver. These transcripts share unexpected characteristics: they are short in length, have long half-lives, are actively translated, and gene ontology analyses revealed that they are enriched in functions in ribosome and oxidative phosphorylation pathways. However, most of these transcripts do not exhibit rhythmicity in poly(A) tail length or steady-state mRNA level, despite Noc’s robust rhythmicity. Therefore, even though the poly(A) tail length dynamics seen between genotypes may not result from direct NOC deadenylase activity, these data suggest that NOC exerts strong effects on physiology through direct and indirect control of target mRNAs. -

The Role of the Mtor Pathway in Developmental Reprogramming Of
THE ROLE OF THE MTOR PATHWAY IN DEVELOPMENTAL REPROGRAMMING OF HEPATIC LIPID METABOLISM AND THE HEPATIC TRANSCRIPTOME AFTER EXPOSURE TO 2,2',4,4'- TETRABROMODIPHENYL ETHER (BDE-47) An Honors Thesis Presented By JOSEPH PAUL MCGAUNN Approved as to style and content by: ________________________________________________________** Alexander Suvorov 05/18/20 10:40 ** Chair ________________________________________________________** Laura V Danai 05/18/20 10:51 ** Committee Member ________________________________________________________** Scott C Garman 05/18/20 10:57 ** Honors Program Director ABSTRACT An emerging hypothesis links the epidemic of metabolic diseases, such as non-alcoholic fatty liver disease (NAFLD) and diabetes with chemical exposures during development. Evidence from our lab and others suggests that developmental exposure to environmentally prevalent flame-retardant BDE47 may permanently reprogram hepatic lipid metabolism, resulting in an NAFLD-like phenotype. Additionally, we have demonstrated that BDE-47 alters the activity of both mTOR complexes (mTORC1 and 2) in hepatocytes. The mTOR pathway integrates environmental information from different signaling pathways, and regulates key cellular functions such as lipid metabolism, innate immunity, and ribosome biogenesis. Thus, we hypothesized that the developmental effects of BDE-47 on liver lipid metabolism are mTOR-dependent. To assess this, we generated mice with liver-specific deletions of mTORC1 or mTORC2 and exposed these mice and their respective controls perinatally to -

Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signifcance and Impact on Clinical Features
Runx1 Gene in Acute Myeloid Leukemia: Expression Level Signicance and Impact on Clinical Features Fadwa Said Abdelazim Mohamed ( [email protected] ) Cairo University Roxan E Shak National Cancer Institute, Cairo University Naglaa M. Hassan National Cancer Institute, Cairo University Research article Keywords: Acute myeloid leukemia, RUNX1, mutation, gene expression, Egypt Posted Date: April 7th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-20926/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/20 Abstract Background Acute myeloid leukemia represents the highest percentage of all adult acute leukemia variants. Runt-related transcription factor1 (RUNX1), a transcription factor with a known tumor suppressor function was recently reported as a tumor promotor in AML. We investigated the role of RUNX1 geneexpression levelin Egyptian AML patients and delineateits clinical signicance. Methods This study recruited 91 AML patients that were recently diagnosed at our hospital with 14 healthy age- and sex-matched donors of bone marrow transplantation unit. We measured RUNX1 gene expression level using reverse transcription–quantitative polymerase chain reaction. Results We found that RUNX1 gene expression level was signicantly higher compared to the control group (p < 0.001). Patients with FLT3 mutations had the higher expression level of RUNX1 (p=0.023). The male patients expressed signicantly higher level of RUNX1 (p=0.046). Conclusion The RUNX1 gene expression may serve as a diagnostic marker in Egyptian AML patients. Its relation to FLT3 may give clue that patients carrying this mutation may benet fromnew treatments that target RUNX1 in the future. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
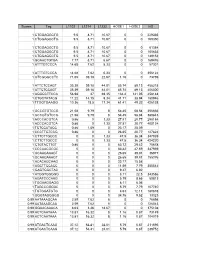
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

(12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer Et Al
USOO8440393B2 (12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer et al. (45) Date of Patent: May 14, 2013 (54) PRO-ANGIOGENIC GENES IN OVARIAN OTHER PUBLICATIONS TUMORENDOTHELIAL CELL, SOLATES Boyd (The Basic Science of Oncology, 1992, McGraw-Hill, Inc., p. (75) Inventors: Michael J. Birrer, Mt. Airy, MD (US); 379). Tomas A. Bonome, Washington, DC Tockman et al. (Cancer Res., 1992, 52:2711s-2718s).* (US); Anil Sood, Pearland, TX (US); Pritzker (Clinical Chemistry, 2002, 48: 1147-1150).* Chunhua Lu, Missouri City, TX (US) Benedict et al. (J. Exp. Medicine, 2001, 193(1) 89-99).* Jiang et al. (J. Biol. Chem., 2003, 278(7) 4763-4769).* (73) Assignees: The United States of America as Matsushita et al. (FEBS Letters, 1999, vol. 443, pp. 348-352).* Represented by the Secretary of the Singh et al. (Glycobiology, 2001, vol. 11, pp. 587-592).* Department of Health and Human Abbosh et al. (Cancer Res. Jun. 1, 2006 66:5582-55.91 and Supple Services, Washington, DC (US); The mental Figs. S1-S7).* University of MD Anderson Cancer Zhai et al. (Chinese General Practice Aug. 2008, 11(8A): 1366 Center, Houston, TX (US) 1367).* Lu et al. (Cancer Res. Feb. 15, 2007, 64(4): 1757-1768).* (*) Notice: Subject to any disclaimer, the term of this Bagnato et al., “Activation of Mitogenic Signaling by Endothelin 1 in patent is extended or adjusted under 35 Ovarian Carcinoma Cells', Cancer Research, vol. 57, pp. 1306-1311, U.S.C. 154(b) by 194 days. 1997. Bouras et al., “Stanniocalcin 2 is an Estrogen-responsive Gene (21) Appl. -

Dissecting Differential Gene Expression Within the Circadian Neuronal Circuit of Drosophila
ART ic LE S Dissecting differential gene expression within the circadian neuronal circuit of Drosophila Emi Nagoshi1,2,4, Ken Sugino2, Ela Kula1,2,4, Etsuko Okazaki3, Taro Tachibana3, Sacha Nelson2 & Michael Rosbash1,2 Behavioral circadian rhythms are controlled by a neuronal circuit consisting of diverse neuronal subgroups. To understand the molecular mechanisms underlying the roles of neuronal subgroups within the Drosophila circadian circuit, we used cell-type specific gene-expression profiling and identified a large number of genes specifically expressed in all clock neurons or in two important subgroups. Moreover, we identified and characterized two circadian genes, which are expressed specifically in subsets of clock cells and affect different aspects of rhythms. The transcription factor Fer2 is expressed in ventral lateral neurons; it is required for the specification of lateral neurons and therefore their ability to drive locomotor rhythms. The Drosophila melanogaster homolog of the vertebrate circadian gene nocturnin is expressed in a subset of dorsal neurons and mediates the circadian light response. The approach should also enable the molecular dissection of many different Drosophila neuronal circuits. A central goal of neurobiology is to understand animal behavior by the functional diversity of clock neurons and how they interact to understanding neuronal circuits. Deciphering how they function and form functional circuits remain largely unknown. thereby generate behavior requires identification of neuronal subtypes From a molecular standpoint, circadian rhythms are generated within a circuit, their organization and connections, as well as the by cell-autonomous molecular clocks present within many cells and contributions of individual cells to overall circuit output. Circadian tissues. -

An Optimized Streptavidin-Binding RNA Aptamer for Purification Of
Published online 23 October 2013 Nucleic Acids Research, 2014, Vol. 42, No. 2 e13 doi:10.1093/nar/gkt956 An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins Kathrin Leppek1,2,3 and Georg Stoecklin1,2,3,* 1Helmholtz Junior Research Group Posttranscriptional Control of Gene Expression, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany, 2Zentrum fu¨ r Molekulare Biologie der Universita¨ t Heidelberg (ZMBH), Im Neuenheimer Feld 282, 69120 Heidelberg, Germany and 3DKFZ-ZMBH Downloaded from https://academic.oup.com/nar/article-abstract/42/2/e13/1030103 by guest on 11 March 2019 Alliance Received June 19, 2013; Revised September 6, 2013; Accepted September 27, 2013 ABSTRACT degradation (1,2). The fate of any given mRNA is essen- tially determined by the messenger ribonucleoprotein Determining the composition of messenger (mRNP) complex, i.e. the ensemble of proteins and regu- ribonucleoprotein (mRNP) particles is essential for latory RNAs an mRNA interacts with. Although decades a comprehensive understanding of the complex of research have uncovered specific functions of numerous mechanisms underlying mRNA regulation, but is RNA-binding proteins (RNA-BPs), surprisingly little is technically challenging. Here we present an RNA- known about the composition, heterogeneity and based method to identify RNP components using a dynamics of whole mRNPs. modified streptavidin (SA)-binding RNA aptamer There are essentially two strategies to explore the termed S1m. By optimizing the RNA aptamer S1 in content of mRNPs. On one hand, protein-based structure and repeat conformation, we improved its approaches rely on the purification of a particular RNA- affinity for SA and found a 4-fold repeat of S1m BP by immunoprecipitation together with its associated mRNAs, whose identity can then be determined by (4ÂS1m) to be more efficient than the established cDNA cloning, microarray analysis (RIP-Chip) or deep MS2 and PP7 systems from bacteriophages. -

The Metabolites NADP+ and NADPH Are the Targets of the Circadian Protein Nocturnin (Curled)
ARTICLE https://doi.org/10.1038/s41467-019-10125-z OPEN The metabolites NADP+ and NADPH are the targets of the circadian protein Nocturnin (Curled) Michael A. Estrella 1,4, Jin Du 1,4, Li Chen2,3,4, Sneha Rath1, Eliza Prangley1, Alisha Chitrakar1, Tsutomu Aoki1, Paul Schedl1, Joshua Rabinowitz2,3 & Alexei Korennykh1 Nocturnin (NOCT) is a rhythmically expressed protein that regulates metabolism under the control of circadian clock. It has been proposed that NOCT deadenylates and regulates 1234567890():,; metabolic enzyme mRNAs. However, in contrast to other deadenylases, purified NOCT lacks the deadenylase activity. To identify the substrate of NOCT, we conducted a mass spec- trometry screen and report that NOCT specifically and directly converts the dinucleotide NADP+ into NAD+ and NADPH into NADH. Further, we demonstrate that the Drosophila NOCT ortholog, Curled, has the same enzymatic activity. We obtained the 2.7 Å crystal structure of the human NOCT•NADPH complex, which revealed that NOCT recognizes the chemically unique ribose-phosphate backbone of the metabolite, placing the 2′-terminal phosphate productively for removal. We provide evidence for NOCT targeting to mito- chondria and propose that NADP(H) regulation, which takes place at least in part in mito- chondria, establishes the molecular link between circadian clock and metabolism. 1 216 Schultz Laboratory, Department of Molecular Biology, Princeton, NJ 08544, USA. 2 285 Frick Laboratory, Department of Chemistry, Princeton, NJ 08544, USA. 3 Lewis-Sigler Institute for Integrative Genomics, Princeton, NJ 08544, USA. 4These authors contributed equally: Michael A. Estrella, Jin Du, Li Chen. Correspondence and requests for materials should be addressed to J.R. -

(12) Patent Application Publication (10) Pub. No.: US 2013/0274315 A1 Birrer Et Al
US 201302743 15A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0274315 A1 Birrer et al. (43) Pub. Date: Oct. 17, 2013 (54) PRO-ANGIOGENC GENES IN OVARAN (60) Provisional application No. 60/901,455, filed on Feb. TUMORENDOTHELIAL CELL, SOLATES 14, 2007. (71) Applicants: The University of Texas MD Anderson Publication Classification Cancer Center, Houston, TX (US); The Government of the U.S.A as (51) Int. Cl. represented by the Secretary of the CI2O I/68 (2006.01) Department of He, Rockville, MD (US) (52) U.S. Cl. CPC .................................... CI2O I/6886 (2013.01) (72) Inventors: Michael J. Birrer, Mt. Airy, MD (US); USPC ............... 514/44A: 435/6.12: 506/9: 435/7.1 Tomas A. Bonome, Washington, DC (US); Anil Sood, Pearland, TX (US); (57) ABSTRACT Chunhua Lu, Missouri City, TX (US) A gene profiling signature for ovarian tumor endothelial cells is disclosed herein. The gene signature can be used to diag nosis or prognosis an ovarian tumor, identify agents to treat an (21) Appl. No.: 13/863,219 ovariantumor, to predict the metastatic potential of an ovarian tumor and to determine the effectiveness of ovarian tumor (22) Filed: Apr. 15, 2013 treatments. Thus, methods are provided for identifying agents that can be used to treat ovarian cancer, for determining the effectiveness of an ovarian tumor treatment, or to diagnose or Related U.S. Application Data prognose an ovarian tumor. Methods of treatment are also (60) Division of application No. 12/541,729, filed on Aug. disclosed which include administering a composition that 14, 2009, now Pat. -

Agricultural University of Athens
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΩΝ ΖΩΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΖΩΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΝΙΚΗΣ ΚΑΙ ΕΙΔΙΚΗΣ ΖΩΟΤΕΧΝΙΑΣ ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ ΑΘΗΝΑ 2020 ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Genome-wide association analysis and gene network analysis for (re)production traits in commercial broilers ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ Τριμελής Επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Επταμελής εξεταστική επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Πηνελόπη Μπεμπέλη (Καθ. ΓΠΑ) Δημήτριος Βλαχάκης (Επ. Καθ. ΓΠΑ) Ευάγγελος Ζωίδης (Επ.Καθ. ΓΠΑ) Γεώργιος Θεοδώρου (Επ.Καθ. ΓΠΑ) 2 Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Περίληψη Σκοπός της παρούσας διδακτορικής διατριβής ήταν ο εντοπισμός γενετικών δεικτών και υποψηφίων γονιδίων που εμπλέκονται στο γενετικό έλεγχο δύο τυπικών πολυγονιδιακών ιδιοτήτων σε κρεοπαραγωγικά ορνίθια. Μία ιδιότητα σχετίζεται με την ανάπτυξη (σωματικό βάρος στις 35 ημέρες, ΣΒ) και η άλλη με την αναπαραγωγική -

Characterization of Molecular Targets for Differential Regulation of the Type I and III Interferon Induction and Signalling Pathways by Rotavirus NSP1
Characterization of molecular targets for differential regulation of the type I and III interferon induction and signalling pathways by rotavirus NSP1 Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy by Gennaro Iaconis September 2018 Table of contents TABLE OF CONTENTS ............................................................................................................................... 1 LIST OF FIGURES ...................................................................................................................................... 6 LIST OF TABLES ........................................................................................................................................ 8 DECLARATION ....................................................................................................................................... 10 ABSTRACT ............................................................................................................................................. 11 1 INTRODUCTION ........................................................................................................................... 12 1.1 Rotavirus ................................................................................................................. 12 1.1.1 Historical Background ......................................................................................... 12 1.1.2 Classification ......................................................................................................