Detection of Metallothionein 1G As a Methylated Tumor Suppressor Gene in Human Hepatocellular Carcinoma Using a Novel Method of Double Combination Array Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metallothionein Monoclonal Antibody, Clone N11-G
Metallothionein monoclonal antibody, clone N11-G Catalog # : MAB9787 規格 : [ 50 uL ] List All Specification Application Image Product Rabbit monoclonal antibody raised against synthetic peptide of MT1A, Western Blot (Recombinant protein) Description: MT1B, MT1E, MT1F, MT1G, MT1H, MT1IP, MT1L, MT1M, MT2A. Immunogen: A synthetic peptide corresponding to N-terminus of human MT1A, MT1B, MT1E, MT1F, MT1G, MT1H, MT1IP, MT1L, MT1M, MT2A. Host: Rabbit enlarge Reactivity: Human, Mouse Immunoprecipitation Form: Liquid Enzyme-linked Immunoabsorbent Assay Recommend Western Blot (1:1000) Usage: ELISA (1:5000-1:10000) The optimal working dilution should be determined by the end user. Storage Buffer: In 20 mM Tris-HCl, pH 8.0 (10 mg/mL BSA, 0.05% sodium azide) Storage Store at -20°C. Instruction: Note: This product contains sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only. Datasheet: Download Applications Western Blot (Recombinant protein) Western blot analysis of recombinant Metallothionein protein with Metallothionein monoclonal antibody, clone N11-G (Cat # MAB9787). Lane 1: 1 ug. Lane 2: 3 ug. Lane 3: 5 ug. Immunoprecipitation Enzyme-linked Immunoabsorbent Assay ASSP5 MT1A MT1B MT1E MT1F MT1G MT1H MT1M MT1L MT1IP Page 1 of 5 2021/6/2 Gene Information Entrez GeneID: 4489 Protein P04731 (Gene ID : 4489);P07438 (Gene ID : 4490);P04732 (Gene ID : Accession#: 4493);P04733 (Gene ID : 4494);P13640 (Gene ID : 4495);P80294 (Gene ID : 4496);P80295 (Gene ID : 4496);Q8N339 (Gene ID : 4499);Q86YX0 (Gene ID : 4490);Q86YX5 -

The Roles of Metallothioneins in Carcinogenesis Manfei Si and Jinghe Lang*
Si and Lang Journal of Hematology & Oncology (2018) 11:107 https://doi.org/10.1186/s13045-018-0645-x REVIEW Open Access The roles of metallothioneins in carcinogenesis Manfei Si and Jinghe Lang* Abstract Metallothioneins (MTs) are small cysteine-rich proteins that play important roles in metal homeostasis and protection against heavy metal toxicity, DNA damage, and oxidative stress. In humans, MTs have four main isoforms (MT1, MT2, MT3, and MT4) that are encoded by genes located on chromosome 16q13. MT1 comprises eight known functional (sub)isoforms (MT1A, MT1B, MT1E, MT1F, MT1G, MT1H, MT1M, and MT1X). Emerging evidence shows that MTs play a pivotal role in tumor formation, progression, and drug resistance. However, the expression of MTs is not universal in all human tumors and may depend on the type and differentiation status of tumors, as well as other environmental stimuli or gene mutations. More importantly, the differential expression of particular MT isoforms can be utilized for tumor diagnosis and therapy. This review summarizes the recent knowledge on the functions and mechanisms of MTs in carcinogenesis and describes the differential expression and regulation of MT isoforms in various malignant tumors. The roles of MTs in tumor growth, differentiation, angiogenesis, metastasis, microenvironment remodeling, immune escape, and drug resistance are also discussed. Finally, this review highlights the potential of MTs as biomarkers for cancer diagnosis and prognosis and introduces some current applications of targeting MT isoforms in cancer therapy. The knowledge on the MTs may provide new insights for treating cancer and bring hope for the elimination of cancer. Keywords: Metallothionein, Metal homeostasis, Cancer, Carcinogenesis, Biomarker Background [6–10]. -

ID AKI Vs Control Fold Change P Value Symbol Entrez Gene Name *In
ID AKI vs control P value Symbol Entrez Gene Name *In case of multiple probesets per gene, one with the highest fold change was selected. Fold Change 208083_s_at 7.88 0.000932 ITGB6 integrin, beta 6 202376_at 6.12 0.000518 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 1553575_at 5.62 0.0033 MT-ND6 NADH dehydrogenase, subunit 6 (complex I) 212768_s_at 5.50 0.000896 OLFM4 olfactomedin 4 206157_at 5.26 0.00177 PTX3 pentraxin 3, long 212531_at 4.26 0.00405 LCN2 lipocalin 2 215646_s_at 4.13 0.00408 VCAN versican 202018_s_at 4.12 0.0318 LTF lactotransferrin 203021_at 4.05 0.0129 SLPI secretory leukocyte peptidase inhibitor 222486_s_at 4.03 0.000329 ADAMTS1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 1552439_s_at 3.82 0.000714 MEGF11 multiple EGF-like-domains 11 210602_s_at 3.74 0.000408 CDH6 cadherin 6, type 2, K-cadherin (fetal kidney) 229947_at 3.62 0.00843 PI15 peptidase inhibitor 15 204006_s_at 3.39 0.00241 FCGR3A Fc fragment of IgG, low affinity IIIa, receptor (CD16a) 202238_s_at 3.29 0.00492 NNMT nicotinamide N-methyltransferase 202917_s_at 3.20 0.00369 S100A8 S100 calcium binding protein A8 215223_s_at 3.17 0.000516 SOD2 superoxide dismutase 2, mitochondrial 204627_s_at 3.04 0.00619 ITGB3 integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) 223217_s_at 2.99 0.00397 NFKBIZ nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta 231067_s_at 2.97 0.00681 AKAP12 A kinase (PRKA) anchor protein 12 224917_at 2.94 0.00256 VMP1/ mir-21likely ortholog -

Changes in Gene Expression by Trabecular Meshwork Cells in Response to Mechanical Stretching
Changes in Gene Expression by Trabecular Meshwork Cells in Response to Mechanical Stretching Vasavi Vittal, Anastasia Rose, Kate E. Gregory, Mary J. Kelley, and Ted S. Acott PURPOSE. Trabecular meshwork (TM) cells appear to sense to 5% of people exhibit pathologic elevations in IOP with changes in intraocular pressure (IOP) as mechanical stretching. subsequent optic nerve damage, even at advanced ages.1,2 We In response, they make homeostatic corrections in the aqueous have hypothesized that TM cells can adjust outflow resistance humor outflow resistance, partially by increasing extracellular over a timescale of hours to days by modulating trabecular ECM matrix (ECM) turnover initiated by the matrix metalloprotein- turnover and subsequent biosynthetic replacement.3–6 Manip- ases. To understand this homeostatic adjustment process fur- ulation of the trabecular activity of a family of ECM turnover ther, studies were conducted to evaluate changes in TM gene enzymes, the matrix metalloproteinases (MMPs), reversibly expression that occur in response to mechanical stretching. modulates outflow facility.7 Inhibition of the endogenous ECM METHODS. Porcine TM cells were subjected to sustained me- turnover, which is initiated by these MMPs, increases the chanical stretching, and RNA was isolated after 12, 24, or 48 outflow resistance. Therefore, ongoing ECM turnover must be hours. Changes in gene expression were evaluated with mi- necessary for homeostatic maintenance of the IOP. In addition, croarrays containing approximately 8000 cDNAs. Select mRNA laser trabeculoplasty, a common treatment for glaucoma, ap- changes were then compared by quantitative reverse transcrip- pears to owe its efficacy to producing relatively sustained tion–polymerase chain reaction (qRT-PCR). -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein -

Boergenomeres2001.Pdf
Letter Identification and Classification of Differentially Expressed Genes in Renal Cell Carcinoma by Expression Profiling on a Global Human 31,500-Element cDNA Array Judith M. Boer,1,5 Wolfgang K. Huber,1 Holger Sültmann,1 Friederike Wilmer,1 Anja von Heydebreck,2,6 Stefan Haas,1,2 Bernhard Korn,1,4 Bastian Gunawan,3 Andreas Vente,4 Laszlo Füzesi,3 Martin Vingron,2,6 and Annemarie Poustka1,7 Departments of 1Molecular Genome Analysis and 2Theoretical Bioinformatics, German Cancer Research Center, Heidelberg, Germany; 3Institute of Pathology, Georg-August-Universita¨t, Go¨ttingen, Germany; 4Resource Center Primary Database, Berlin/Heidelberg, Germany We investigated the changes in gene expression accompanying the development and progression of kidney cancer by use of 31,500-element complementary DNA arrays. We measured expression profiles for paired neoplastic and noncancerous renal epithelium samples from 37 individuals. Using an experimental design optimized for factoring out technological and biological noise, and an adapted statistical test, we found 1738 differentially expressed cDNAs with an expected number of six false positives. Functional annotation of these genes provided views of the changes in the activities of specific biological pathways in renal cancer. Cell adhesion, signal transduction, and nucleotide metabolism were among the biological processes with a large proportion of genes overexpressed in renal cell carcinoma. Down-regulated pathways in the kidney tumor cells included small molecule transport, ion homeostasis, and oxygen and radical metabolism. Our expression profiling data uncovered gene expression changes shared with other epithelial tumors, as well as a unique signature for renal cell carcinoma. [Expression data for the differentially expressed cDNAs are available as a Web supplement at http://www.dkfz- heidelberg.de/abt0840/whuber/rcc. -

The Journal of Toxicological Sciences (J
The Journal of Toxicological Sciences (J. Toxicol. Sci.) 147 Vol.41, No.1, 147-153, 2016 Original Article Upregulations of metallothionein gene expressions and tolerance to heavy metal toxicity by three dimensional cultivation of HepG2 cells on VECELL 3-D inserts Takashi Kubo1,2, Yukie Kuroda1, Shinichiro Horiuchi1, Su-Ryang Kim1, Yuko Sekino1 and Seiichi Ishida1 1Division of Pharmacology, National Institute of Health Sciences, 1-18-1 Kamiyoga, Setagaya-ku, Tokyo 158-8501, Japan 2Present address: Division of Translationl Research, Exploratory Oncology Research & Clinical Trial Center, National Cancer Center, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan (Received October 9, 2015; Accepted December 6, 2015) ABSTRACT — The VECELL 3-D insert is a new culture scaffold consisting of collagen-coated ePTFE (expanded polytetrafluoroethylene) mesh. We analyzed the effects of VECELL 3-D inserts on the func- tionality of HepG2, a human hepatocellular carcinoma cell line. HepG2 cells cultured on VECELL 3-D inserts maintained a round shape, while those cultured on a standard culture plate or collagen-coated cell culture plate showed a flattened and cubic epithelial-like shape. HepG2 cells cultured on VECELL 3-D inserts had showed upregulated expression of metallothionein genes and in turn a higher tolerance to toxicity induced by heavy metals. These results suggest that HepG2 cell functions were changed by the cell morphology that is induced by culturing on a VECELL 3-D insert. Key words: VECELL 3-D insert, HepG2 cell, 3-D culture, Metallothionein, Heavy metal toxicity, Global gene expression analysis INTRODUCTION sion analysis revealed that the changes in expression of microtubule molecules might be responsible for these Several studies have shown changes in cell functions functional changes. -

Increased Interleukin-11 and Stress-Related Gene Expression in Human Endothelial and Bronchial Epithelial Cells Exposed to Silver Nanoparticles
biomolecules Article Increased Interleukin-11 and Stress-Related Gene Expression in Human Endothelial and Bronchial Epithelial Cells Exposed to Silver Nanoparticles Jiyoung Jang 1,2,3 , Sun Park 4,5,* and In-Hong Choi 1,2,* 1 Department of Microbiology, Yonsei University College of Medicine, Seoul 03722, Korea; [email protected] 2 Institute for Immunology and Immunological Diseases, Yonsei University College of Medicine, Seoul 03722, Korea 3 Humidifier Disinfectant Health Center, National Institute of Environmental Research, Incheon 22689, Korea 4 Department of Biomedical Sciences, The Graduate School, Ajou University, Suwon 16499, Korea 5 Department of Microbiology, Ajou University School of Medicine, Suwon 16499, Korea * Correspondence: [email protected] (S.P.); [email protected] (I.-H.C.); Tel.: +82-31-219-5071 (S.P.); +82-2-2228-1821 (I.-H.C.) Abstract: This article aimed to identify and distinguish the various responses to silver nanoparticles (NPs) of endothelial and epithelial cells. We also assessed the significantly increased gene expression levels, as shown by microarray analysis. We evaluated the median lethal dose of NPs in each cell line and found that each value was different. We also confirmed the toxicity of 5 nm silver NPs. Meanwhile, cell death was not observed in cells exposed to 100 nm silver NPs at a high concentration. We verified that 5 nm silver NPs affected the variation in gene expression in cells through microarray analysis and observed a noticeable increase in interleukin (IL)-8 and IL-11 gene expression in early Citation: Jang, J.; Park, S.; Choi, I.-H. stages. This study showed noticeable variation in the expression of oxidative stress-related genes Increased Interleukin-11 and in early stages. -

Metallothionein 1G Promotes the Differentiation of HT-29 Human Colorectal Cancer Cells
ONCOLOGY REPORTS 37: 2633-2651, 2017 Metallothionein 1G promotes the differentiation of HT-29 human colorectal cancer cells JUAN MartÍN ARRIAGA1,2, ALICIA INÉS BRAVO3, JOSÉ MORDOH1,2,4 and MICHELE BIANCHINI2 1Cancerology Laboratory, Leloir Institute, IIBBA-CONICET, Buenos Aires 1405; 2Center for Oncology Research, Cancer Foundation (CIO-FUCA), Buenos Aires 1426; 3Acute Interzonal General Hospiutal ‘Eva Perón’, Buenos Aires 1650; 4Alexander Fleming Institute, Buenos Aires 1180, Argentina Received September 27, 2016; Accepted November 16, 2016 DOI: 10.3892/or.2017.5547 Abstract. Metallothioneins (MTs) are a family of low- CRC tumors, and implicate MTs and zinc signaling as new molecular-weight, cysteine-rich proteins involved in zinc players in colorectal differentiation. This further contributes to and redox metabolism, that are epigenetically downregulated the hypothesis that re-induction of MTs may have therapeutic during colorectal cancer (CRC) progression, but may be value by diminishing the aggressiveness of CRC tumors. re-induced with a variety of agents. Since loss of MT expres- sion is associated with a worse prognosis, in the present study Introduction we investigated the effects of overexpression of the most significantly downregulated isoform in CRC, namely MT1G, Colorectal cancer (CRC) is the third most commonly on the HT-29 cell line. Overexpression of MT1G resulted in diagnosed cancer worldwide, having a mortality rate near xenograft tumors with an aberrant morphology, character- 50% (1). Recent studies have shown that these tumors retain ized by an evident increase in mucin-containing cells that multilineage differentiation processes similar to those of were identified as goblet cells under electron microscopy. the normal intestinal epithelium, mainly the goblet cell and Immunohistochemical detection of CDX2 and cytokeratin 20 enterocyte lineages (2). -

Identification of the Collagen Type 1 Alpha 1 Gene (COL1A1)
Hayashi et al. BMC Cancer 2014, 14:108 http://www.biomedcentral.com/1471-2407/14/108 RESEARCH ARTICLE Open Access Identification of the collagen type 1 alpha 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma Masamichi Hayashi, Shuji Nomoto*, Mitsuhiro Hishida, Yoshikuni Inokawa, Mitsuro Kanda, Yukiyasu Okamura, Yoko Nishikawa, Chie Tanaka, Daisuke Kobayashi, Suguru Yamada, Goro Nakayama, Tsutomu Fujii, Hiroyuki Sugimoto, Masahiko Koike, Michitaka Fujiwara, Shin Takeda and Yasuhiro Kodera Abstract Background: Hepatocellular carcinoma (HCC) is one of the major causes of cancer-related death especially among Asian and African populations. It is urgent that we identify carcinogenesis-related genes to establish an innovative treatment strategy for this disease. Methods: Triple-combination array analysis was performed using one pair each of HCC and noncancerous liver samples from a 68-year-old woman. This analysis consists of expression array, single nucleotide polymorphism array and methylation array. The gene encoding collagen type 1 alpha 1 (COL1A1) was identified and verified using HCC cell lines and 48 tissues from patients with primary HCC. Results: Expression array revealed that COL1A1 gene expression was markedly decreased in tumor tissues (log2 ratio –1.1). The single nucleotide polymorphism array showed no chromosomal deletion in the locus of COL1A1. Importantly, the methylation value in the tumor tissue was higher (0.557) than that of the adjacent liver tissue (0.008). We verified that expression of this gene was suppressed by promoter methylation. Reactivation of COL1A1 expression by 5-aza-2′-deoxycytidine treatment was seen in HCC cell lines, and sequence analysis identified methylated CpG sites in the COL1A1 promoter region. -
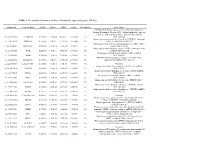
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137. -

Metallothionein 1G Acts As an Oncosupressor in Papillary Thyroid
Laboratory Investigation (2008) 88, 474–481 & 2008 USCAP, Inc All rights reserved 0023-6837/08 $30.00 Metallothionein 1G acts as an oncosupressor in papillary thyroid carcinoma Cristina Ferrario1,2, Paola Lavagni1, Manuela Gariboldi1,2, Claudia Miranda1, Marco Losa3, Loredana Cleris4, Franca Formelli4, Silvana Pilotti3, Marco A Pierotti1,2,5 and Angela Greco1 The molecular pathogenesis of tumors arising from the thyroid follicular epithelial cells, including papillary (PTC) and follicular thyroid carcinoma (FTC), is only partially understood, and the role of tumor suppressor genes has not yet been assessed. The metallothionein (MT) gene family encodes a class of metal-binding proteins involved in several cellular processes, and their expression is often deregulated in human tumors. Recently, downregulation of MT gene expression in PTC has been reported, suggesting a possible oncosuppressor role of this gene family in the pathogenesis of thyroid tumors. To further explore this possibility, we performed expression and functional studies. Analysis of microarray data of thyroid tumors of different histologic types showed that several MT genes were downregulated with respect to normal tissue. The microarray data were corroborated by quantitative PCR experiments, showing downregulation of MTs in PTC and FTC, but to a greater extent in papillary carcinoma. The expression of MTs was also investigated at the protein level by immunohistochemistry; the results were consistent with the microarray data, showing general downregulation in tumor samples, which was more evident in PTC. The functional consequence of MT downregulation was addressed employing an experimental model made of the PTC-derived K1 cell line in which MT1G expression is repressed by promoter methylation.