The Expanding Toolbox of In Vivo Bioluminescent Imaging
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Where Is That Light Coming From?
2004 Deep-Scope Expedition Where is That Light Coming From? FOCUS TEACHING TIME Bioluminescence One 45-minute class period, plus time for student research GRADE LEVEL 9-12 (Chemistry & Life Science) SEATING ARRANGEMENT Classroom style or groups of 3-4 students FOCUS QUESTION What is the basic chemistry responsible for bio- MAXIMUM NUMBER OF STUDENTS luminescence, and how does this process benefit 30 deep-sea organisms? KEY WORDS LEARNING OBJECTIVES Chemiluminescence Students will be able to explain the role of lucif- Bioluminescence erins, luciferases, and co-factors in biolumines- Fluorescence cence, and the general sequence of the light-emit- Phosphorescence ting process. Luciferin Luciferase Students will be able to discuss the major types of Photoprotein luciferins found in marine organisms. Counter-illumination Lux operon Students will be able to define the “lux operon?” BACKGROUND INFORMATION Students will be able to discuss at least three Deep-sea explorers face many challenges: ways that bioluminescence may benefit deep-sea extreme heat and cold, high pressures, and organisms, and give an example of at least one almost total darkness. The absence of light poses organism that actually receives each of the ben- particular challenges to scientists who want to efits discussed. study organisms that inhabit the deep ocean envi- ronment. Even though deep-diving submersibles MATERIALS carry bright lights, simply turning these lights on ❑ None creates another set of problems: at least some mobile organisms are likely to move away from -

William D. Mcelroy Papers
http://oac.cdlib.org/findaid/ark:/13030/kt6489r0v5 No online items William D. McElroy Papers Special Collections & Archives, UC San Diego Special Collections & Archives, UC San Diego Copyright 2005 9500 Gilman Drive La Jolla 92093-0175 [email protected] URL: http://libraries.ucsd.edu/collections/sca/index.html William D. McElroy Papers MSS 0483 1 Descriptive Summary Languages: English Contributing Institution: Special Collections & Archives, UC San Diego 9500 Gilman Drive La Jolla 92093-0175 Title: William D. McElroy Papers Identifier/Call Number: MSS 0483 Physical Description: 10.4 Linear feet(19 archives boxes, 11 oversize folders, 6 art bin items) Date (inclusive): 1944-1999 Abstract: Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. Scope and Content of Collection Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. McElroy's significant contributions to biology include isolating and crystallizing the compounds that enable firefly luminescence and for his subsequent research into bacterial bioluminescence. He also wrote, spoke, and worked on problems in areas of environment, pollution, food production, science education, and international science. The papers largely document McElroy's scientific research and include correspondence with the scientific community, various biographical materials including awards and photographs, his trip to China in 1979, writings and reprints related to biochemical and scientific investigation, research materials on bioluminescence, teaching materials, and speeches given both as chancellor and as director of the National Science Foundation. -

Safety Data Sheet
G-Biosciences, St Louis, MO, USA | 1-800-628-7730 | 1-314-991-6034 | [email protected] A Geno Technology, Inc. (USA) brand name Safety Data Sheet Cat. # RC-227 D-Luciferin Firefly, FREE ACID Size: 0.25g think proteins! think G-Biosciences! www.GBiosciences.com D-Luciferin Firefly, potassium salt Safety Data Sheet according to Federal Register / Vol. 77, No. 58 / Monday, March 26, 2012 / Rules and Regulations Date of issue: 05/06/2013 Revision date: 05/11/2017 Version: 7.1 SECTION 1: Identification 1.1. Identification Product form : Substance Trade name : D-Luciferin Firefly, potassium salt CAS-No. : 2591-17-5 Product code : 006A Formula : C11H8N2O3S2 Synonyms : (4S)-4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid / (S)-2-(6-hydroxy-2- benzothiazolyl)-2-thiazoline-4-carboxylic acid / (S)-4,5-dihydro-2-(6-hydroxybenzothiazol-2- yl)thiazole-4-carboxylic acid / 2-(6-hydroxy-2-benzothiazolyl)-2-thiazoline-4-carcoxylic acid, (S)- / 2-(6-hydroxybenzothiazol-2-yl)-2-thiazoline-4-carboxylic acid / 4,5-dihydro-2-(6-hydroxy-2- benzorhiazolyl)-4-thiazolecarboxylic acid, (4S)- / 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4- thiazolecarboxylic acid / 4-Thiazolecarboxylic acid, 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-, (S)- / D-(-)-luciferin / D-luciferin / firefly luciferin / liciferin, D-(-)- / luciferin / luciferin, D- Other means of identification : D-Luciferin Firefly, free acid 4,5-Dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid, ST50405784, Luciferin, CHEBI:17165 BIG No : 48631 1.2. Recommended use and restrictions on use Use of the substance/mixture : Luciferin is a common bioluminescent reporter used for in-vivo imaging of the expression of the luc marker gene . -

Review on Bioluminescence Imaging
DOI: 10.37421/apn.2020.05.170 Review Article Journal of Volume 12:7, 2021 Nuclear Medicine & Radiation Therapy ISSN: 2155-9619 Open Access Review on Bioluminescence Imaging Cleodora Yen* Department of Radiology, Yale University, United States Bioluminescence imaging (BLI) is an innovation created over the previous decade The strategy expects luciferin to be added to the circulatory system, which conveys it that takes into consideration the noninvasive investigation of continuous organic cycles. to cells all through the body. At the point when luciferin arrives at cells that have been As of late, bioluminescence tomography (BLT) has become conceivable and a few adjusted to convey the firefly quality, those cells produce light [2]. frameworks have opened up. In 2011, PerkinElmer gained perhaps the most famous lines of optical imaging frameworks with bioluminescence from Caliper Life Sciences. The BLT opposite issue of 3D remaking of the dispersion of bioluminescent atoms Bioluminescence is the cycle of light outflow in living life forms. Bioluminescence imaging from information estimated on the creature surface is innately poorly presented. The uses local light outflow from one of a few organic entities which bioluminesce. The three main little creature study utilizing BLT was directed by scientists at the University of principle sources are the North American firefly, the ocean pansy (and related marine Southern California, Los Angeles, USA in 2005. Following this turn of events, many organic entities), and microbes like Photorhabdus luminescens and Vibrio fischeri. The exploration bunches in USA and China have fabricated frameworks that empower BLT. DNA encoding the iridescent protein is fused into the lab creature either by means of Mustard plants have had the quality that makes fireflies' tails shine added to them so the a viral vector or by making a transgenic creature. -

Lightning Bugs
GENERAL I ARTICLE Lightning Bugs B Gajendra Babu and M Kannan Bioluminescence is the phenomenon of light emission by B Gajendra Babu and living organisms. This is well exhibited in many insects, M Kannan are PhD Scholars in the Depart and best understood in fireflies. Bioluminescence is the ment of Agricultural result of chemical reactions primarily involving luciferin, Entomology, Tamil Nadu luciferase and oxygen. Luciferin is a heat-resistant sub Agricultural University, strate and the source of light; luciferase, an enzyme, is the Coimbatore. trigger, and oxygen is the fuel. Luminescing insects utilize light as a mating signal, to attract their prey, or to defend themselves from enemies. This biological phenomenon has been exploited in space and medical research, insect pest management, and is also a useful tool in biotechnology. Bioluminescence is the ability of certain animals to produce light, a phenomenon primarily seen in marine organisms. It is the predominant source of light in deep oceans. The light production is the result of chemical reactions and hence it is also called 'chemiluminescence'. Bioluminescence is exhibited by bacteria, fungi, jellyfish, insects, algae, fish, clams, snails, crus taceans, etc. Bioluminescent bacteria have been found in ma rine; coastal and terrestrial environments. Some fungi can also emit light. Luminescent fungi such as Armillaria mellea and Mycena spp. produce a continuous (non-pulsing) light in their fruiting bodies and mycelium. It is believed that biolumines cent fungi use their light to attract insects that will spread the fungal spores, thus enhancing their reproduction. Some nema todes are luminescent due to the presence of symbiotic bacteria associated with them. -

Bioluminescence Is Produced by a Firefly-Like Luciferase but an Entirely
www.nature.com/scientificreports OPEN New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a Received: 8 November 2017 Accepted: 1 February 2018 frefy-like luciferase but an entirely Published: xx xx xxxx new luciferin Oliver C. Watkins1,2, Miriam L. Sharpe 1, Nigel B. Perry 2 & Kurt L. Krause 1 The New Zealand glowworm, Arachnocampa luminosa, is well-known for displays of blue-green bioluminescence, but details of its bioluminescent chemistry have been elusive. The glowworm is evolutionarily distant from other bioluminescent creatures studied in detail, including the frefy. We have isolated and characterised the molecular components of the glowworm luciferase-luciferin system using chromatography, mass spectrometry and 1H NMR spectroscopy. The purifed luciferase enzyme is in the same protein family as frefy luciferase (31% sequence identity). However, the luciferin substrate of this enzyme is produced from xanthurenic acid and tyrosine, and is entirely diferent to that of the frefy and known luciferins of other glowing creatures. A candidate luciferin structure is proposed, which needs to be confrmed by chemical synthesis and bioluminescence assays. These fndings show that luciferases can evolve independently from the same family of enzymes to produce light using structurally diferent luciferins. Glowworms are found in New Zealand and Australia, and are a major tourist attraction at sites located across both countries. In contrast to luminescent beetles such as the frefy (Coleoptera), whose bioluminescence has been well characterised (reviewed by ref.1), the molecular details of glowworm bioluminescence have remained elusive. Tese glowworms are the larvae of fungus gnats of the genus Arachnocampa, with eight species endemic to Australia and a single species found only in New Zealand2. -

Integration of Circadian and Phototransduction Pathways in the Network Controlling CAB Gene Transcription in Arabidopsis
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 15491–15496, December 1996 Plant Biology Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis (firefly luciferaseyluminescence imagingycircadian rhythmygene expression) ANDREW J. MILLAR* AND STEVE A. KAY National Science Foundation Center for Biological Timing, Department of Biology, University of Virginia, Charlottesville, VA 22903 Communicated by Winslow Briggs, Carnegie Institute of Washington, Stanford, CA, October 15, 1996 (received for review June 24, 1996) ABSTRACT The transcription of CAB genes, encoding the the circadian clock acts negatively on CAB expression, antag- chlorophyll ayb-binding proteins, is rapidly induced in dark- onizing the positive effects of phototransduction pathways grown Arabidopsis seedlings following a light pulse. The during the subjective night. transient induction is followed by several cycles of a circadian The transient induction of CAB expression immediately rhythm. Seedlings transferred to continuous light are known after plants are exposed to light is referred to as the ‘‘acute’’ to exhibit a robust circadian rhythm of CAB expression. The response to light (9, 11). Many circadian-regulated processes precise waveform of CAB expression in light–dark cycles, exhibit acute effects of light: the expression of clock-controlled however, reflects a regulatory network that integrates infor- genes (ccg)inNeurospora is induced by light (12), for example, mation from photoreceptors, from the circadian clock and and the production of melatonin in cultured avian pinealocytes possibly from a developmental program. We have used the is suppressed (13). Current evidence indicates that the circa- luciferase reporter system to investigate CAB expression with dian oscillator is not required for these acute responses: ccg high time resolution. -
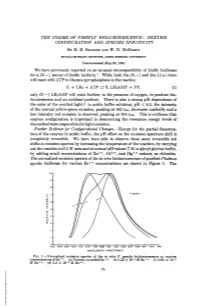
The Colors of Firefly Bioluminescence: Enzyme Configuration and Species Specificity by H
THE COLORS OF FIREFLY BIOLUMINESCENCE: ENZYME CONFIGURATION AND SPECIES SPECIFICITY BY H. H. SELIGER AND W. D. MCELROY MCCOLLUM-PRATT INSTITUTE, JOHNS HOPKINS UNIVERSITY Communicated May 25, 1964 We have previously reported on an unusual stereospecificity of firefly luciferase for a D(-) isomer of firefly luciferin.' While both the D(-) and the L(+) form will react with ATP to liberate pyrophosphate in the reaction E + LH2 + ATP =- E. LH2AMP + PP, (1) only D(-) LH2AMP will react further, in the presence of oxygen, to produce bio- luminescence and an oxidized product. There is also a strong pH dependence of the color of the emitted light;2 in acidic buffer solutions, pH < 6.5, the intensity of the normal yellow-green emission, peaking at 562 ml,, decreases markedly and a low intensity red emission is observed, peaking at 616 miu. This is evidence that enzyme configuration is important in determining the resonance energy levels of the excited state responsible for light emission. Further Evidence for Configurational Changes.-Except for the partial denatura- tion of the enzyme in acidic buffer, the pH effect on the emission spectrum shift is completely reversible. We have been able to observe these same reversible red shifts in emission spectra by increasing the temperature of the reaction, by carrying out the reaction in 0.2 M urea and at normal pH values (7.6) in glycyl glycine buffer, by adding small concentrations of Zn++, Cd++, and Hg++ cations, as chlorides. The normalized emission spectra of the in vitro bioluminescence of purified Photinus pyralis luciferase for various Zn++ concentrations are shown in Figure 1. -

Hidden Diversity in the Brazilian Atlantic Rainforest
www.nature.com/scientificreports Corrected: Author Correction OPEN Hidden diversity in the Brazilian Atlantic rainforest: the discovery of Jurasaidae, a new beetle family (Coleoptera, Elateroidea) with neotenic females Simone Policena Rosa1, Cleide Costa2, Katja Kramp3 & Robin Kundrata4* Beetles are the most species-rich animal radiation and are among the historically most intensively studied insect groups. Consequently, the vast majority of their higher-level taxa had already been described about a century ago. In the 21st century, thus far, only three beetle families have been described de novo based on newly collected material. Here, we report the discovery of a completely new lineage of soft-bodied neotenic beetles from the Brazilian Atlantic rainforest, which is one of the most diverse and also most endangered biomes on the planet. We identifed three species in two genera, which difer in morphology of all life stages and exhibit diferent degrees of neoteny in females. We provide a formal description of this lineage for which we propose the new family Jurasaidae. Molecular phylogeny recovered Jurasaidae within the basal grade in Elateroidea, sister to the well-sclerotized rare click beetles, Cerophytidae. This placement is supported by several larval characters including the modifed mouthparts. The discovery of a new beetle family, which is due to the limited dispersal capability and cryptic lifestyle of its wingless females bound to long-term stable habitats, highlights the importance of the Brazilian Atlantic rainforest as a top priority area for nature conservation. Coleoptera (beetles) is by far the largest insect order by number of described species. Approximately 400,000 species have been described, and many new ones are still frequently being discovered even in regions with histor- ically high collecting activity1. -

Bioluminescence in Insect
Int.J.Curr.Microbiol.App.Sci (2018) 7(3): 187-193 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 7 Number 03 (2018) Journal homepage: http://www.ijcmas.com Review Article https://doi.org/10.20546/ijcmas.2018.703.022 Bioluminescence in Insect I. Yimjenjang Longkumer and Ram Kumar* Department of Entomology, Dr. Rajendra Prasad Central Agricultural University, Pusa, Bihar-848125, India *Corresponding author ABSTRACT Bioluminescence is defined as the emission of light from a living organism K e yw or ds that performs some biological function. Bioluminescence is one of the Fireflies, oldest fields of scientific study almost dating from the first written records Bioluminescence , of the ancient Greeks. This article describes the investigations of insect Luciferin luminescence and the crucial role imparted in the activities of insect. Many Article Info facets of this field are easily accessible for investigation without need for Accepted: advanced technology and so, within the History of Science, investigations 04 February 2018 of bioluminescence played a significant role in the establishment of the Available Online: scientific method, and also were among the many visual phenomena to be 10 March 2018 accounted for in developing a theory of light. Introduction Bioluminescence (BL) serves various purposes, including sexual attraction and When a living organism produces and emits courtship, predation and defense (Hastings and light as a result of a chemical reaction, the Wilson, 1976). This process is suggested to process is known as Bioluminescence - bio have arisen after O2 appearance on Earth at means 'living' in Greek while `lumen means least 30 different times during evolution, as 'light' in Latin. -

Determining the Depth Limit of Bioluminescent Sources in Scattering Media
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.21.044982; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Determining the Depth Limit of Bioluminescent Sources in Scattering Media. Ankit Raghuram1,*, Fan Ye1, Jesse K. Adams1,2, Nathan Shaner3, Jacob T. Robinson1,2,4,5, Ashok Veeraraghavan1,2,6 1 Department of Electrical and Computer Engineering, Rice University, Houston, TX 77005, USA 2 Applied Physics Program, Rice University, Houston, TX 77005, USA 3 Department of Neurosciences, University of California San Diego, La Jolla, CA 92093, USA 4 Department of Neuroscience, Baylor College of Medicine, Houston, TX 77030, USA 5 Department of Bioengineering, Rice University, Houston, TX 77005, USA 6 Department of Computer Science, Rice University, Houston TX 77005, USA * [email protected] Abstract Bioluminescence has several potential advantages compared to fluorescence microscopy for in vivo biological imaging. Because bioluminescence does not require excitation light, imaging can be performed for extended periods of time without phototoxicity or photobleaching, and optical systems can be smaller, simpler, and lighter. Eliminating the need for excitation light may also affect how deeply one can image in scattering biological tissue, but the imaging depth limits for bioluminescence have yet to be reported. Here, we perform a theoretical study of the depth limits of bioluminescence microscopy and find that cellular resolution imaging should be possible at a depth of 5-10 mean free paths (MFPs). This limit is deeper than the depth limit for confocal microscopy and slightly lower than the imaging limit expected for two-photon microscopy under similar conditions. -

Bioluminescence Imaging of DNA Synthetic Phase of Cell Cycle in Living Animals
Bioluminescence Imaging of DNA Synthetic Phase of Cell Cycle in Living Animals Zhi-Hong Chen1,2, Rui-Jun Zhao1,3, Rong-Hui Li1,3, Cui-Ping Guo1,3, Guo-Jun Zhang1,3* 1 Breast Center, Cancer Hospital of Shantou University Medical College, Shantou, People’s Republic of China, 2 Heilongjiang Province Key Laboratory of Cancer Prevention and Treatment, Mudanjiang Medical University, Mudanjiang, People’s Republic of China, 3 Cancer Research Center, Shantou University Medical College, Shantou, People’s Republic of China Abstract Bioluminescence reporter proteins have been widely used in the development of tools for monitoring biological events in living cells. Currently, some assays like flow cytometry analysis are available for studying DNA synthetic phase (S-phase) targeted anti-cancer drug activity in vitro; however, techniques for imaging of in vivo models remain limited. Cyclin A2 is known to promote S-phase entry in mammals. Its expression levels are low during G1-phase, but they increase at the onset of S-phase. Cyclin A2 is degraded during prometaphase by ubiquitin-dependent, proteasome-mediated proteolysis. In this study, we have developed a cyclin A2-luciferase (CYCA-Luc) fusion protein targeted for ubiquitin-proteasome dependent degradation, and have evaluated its utility in screening S-phase targeted anti-cancer drugs. Similar to endogenous cyclin A2, CYCA-Luc accumulates during S-phase and is degraded during G2/M-phase. Using Cdc20 siRNA we have demonstrated that Cdc20 can mediate CYCA-Luc degradation. Moreover, using noninvasive bioluminescent imaging, we demonstrated accumulation of CYCA-Luc in response to 10-hydroxycamptothecin (HCPT), an S-phase targeted anti-cancer drug, in human tumor cells in vivo and in vitro.