A Gene-Anchored Map Position of the Rat Warfarin-Resistance Locus, Rw, and Its Orthologs in Mice and Humans
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic and Genomic Analysis of Hyperlipidemia, Obesity and Diabetes Using (C57BL/6J × TALLYHO/Jngj) F2 Mice
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Nutrition Publications and Other Works Nutrition 12-19-2010 Genetic and genomic analysis of hyperlipidemia, obesity and diabetes using (C57BL/6J × TALLYHO/JngJ) F2 mice Taryn P. Stewart Marshall University Hyoung Y. Kim University of Tennessee - Knoxville, [email protected] Arnold M. Saxton University of Tennessee - Knoxville, [email protected] Jung H. Kim Marshall University Follow this and additional works at: https://trace.tennessee.edu/utk_nutrpubs Part of the Animal Sciences Commons, and the Nutrition Commons Recommended Citation BMC Genomics 2010, 11:713 doi:10.1186/1471-2164-11-713 This Article is brought to you for free and open access by the Nutrition at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Nutrition Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Stewart et al. BMC Genomics 2010, 11:713 http://www.biomedcentral.com/1471-2164/11/713 RESEARCH ARTICLE Open Access Genetic and genomic analysis of hyperlipidemia, obesity and diabetes using (C57BL/6J × TALLYHO/JngJ) F2 mice Taryn P Stewart1, Hyoung Yon Kim2, Arnold M Saxton3, Jung Han Kim1* Abstract Background: Type 2 diabetes (T2D) is the most common form of diabetes in humans and is closely associated with dyslipidemia and obesity that magnifies the mortality and morbidity related to T2D. The genetic contribution to human T2D and related metabolic disorders is evident, and mostly follows polygenic inheritance. The TALLYHO/ JngJ (TH) mice are a polygenic model for T2D characterized by obesity, hyperinsulinemia, impaired glucose uptake and tolerance, hyperlipidemia, and hyperglycemia. -

Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals
animals Review Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals Mohammadreza Mohammadabadi 1 , Farhad Bordbar 1,* , Just Jensen 2 , Min Du 3 and Wei Guo 4 1 Department of Animal Science, Faculty of Agriculture, Shahid Bahonar University of Kerman, Kerman 77951, Iran; [email protected] 2 Center for Quantitative Genetics and Genomics, Aarhus University, 8210 Aarhus, Denmark; [email protected] 3 Washington Center for Muscle Biology, Department of Animal Sciences, Washington State University, Pullman, WA 99163, USA; [email protected] 4 Muscle Biology and Animal Biologics, Animal and Dairy Science, University of Wisconsin-Madison, Madison, WI 53558, USA; [email protected] * Correspondence: [email protected] Simple Summary: Skeletal muscle mass is an important economic trait, and muscle development and growth is a crucial factor to supply enough meat for human consumption. Thus, understanding (candidate) genes regulating skeletal muscle development is crucial for understanding molecular genetic regulation of muscle growth and can be benefit the meat industry toward the goal of in- creasing meat yields. During the past years, significant progress has been made for understanding these mechanisms, and thus, we decided to write a comprehensive review covering regulators and (candidate) genes crucial for muscle development and growth in farm animals. Detection of these genes and factors increases our understanding of muscle growth and development and is a great help for breeders to satisfy demands for meat production on a global scale. Citation: Mohammadabadi, M.; Abstract: Farm-animal species play crucial roles in satisfying demands for meat on a global scale, Bordbar, F.; Jensen, J.; Du, M.; Guo, W. -

Molecular Bases of Equine Polysaccharide Storage Myopathies a Dissertation SUBMITTED to the FACULTY of UNIVERSITY of MINNESOTA
Molecular bases of equine polysaccharide storage myopathies A Dissertation SUBMITTED TO THE FACULTY OF UNIVERSITY OF MINNESOTA BY Raffaella Bertoni Cavalcanti Teixeira IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Molly McCue, James Mickelson April, 2015 © {Raffaella Bertoni Cavalcanti Teixeira, 2015} Acknowledgements First of all I am thankful to Drs McCue and Mickelson for their essential support. Without their superior knowledge and experience I would not have been able to accomplish this work. The author would also like to thank Drs Reed, Rendahl and Valberg for being part of my committee and for their support and help. I would also like to acknowledge Rob Schaefer for all his help with scripts and commands. I have learned a lot from him. I would like to thank my lab mates Sam Beeson, Elaine Norton, Annette McCoy, Felipe Avila, Nichol Schultz and Ann Kemper for their help and friendship. And last, I want to thank my dog Stella for showing me her love during good and bad times. I have met incredible people during this journey and I’m very thankful to each one of them. You have all left a mark on my life forever. i Dedication This thesis is dedicated to my adviser Dr McCue for helping me achieve my goal of completing a PhD. Dr McCue has taught me so many important lessons. I’m a better scientist, clinician and most of all, a better person because of her. Her lessons will stay with me for the rest of my life. I also dedicate this work to my co-adviser, Dr Mickelson, for being supportive, helpful, wise and very patient. -

Gene- and Tissue-Level Interactions in Normal Gastrointestinal Development and Hirschsprung Disease
Gene- and tissue-level interactions in normal gastrointestinal development and Hirschsprung disease Sumantra Chatterjeea,b,1, Priyanka Nandakumara,1, Dallas R. Auera,b, Stacey B. Gabrielc, and Aravinda Chakravartia,b,2 aCenter for Complex Disease Genomics, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205; bCenter for Human Genetics and Genomics, New York University School of Medicine, New York, NY 10016; and cGenomics Platform, Broad Institute of MIT and Harvard, Cambridge, MA 02142 Contributed by Aravinda Chakravarti, November 1, 2019 (sent for review May 21, 2019; reviewed by William J. Pavan and Tatjana Sauka-Spengler) The development of the gut from endodermal tissue to an organ between the longitudinal and circular muscles, and the sub- with multiple distinct structures and functions occurs over a mucosal (Meissner’s) plexus, between the circular muscle and prolonged time during embryonic days E10.5–E14.5 in the mouse. the submucosal layer. The myenteric and submucoal plexuses During this process, one major event is innervation of the gut by provide motor innervation to both muscular layers of the gut, enteric neural crest cells (ENCCs) to establish the enteric nervous and secretomotor innervation of the mucosa nearest the lumen system (ENS). To understand the molecular processes underpinning of the gut, respectively (6). gut and ENS development, we generated RNA-sequencing profiles The many stages of gut development require numerous initiat- from wild-type mouse guts at E10.5, E12.5, and E14.5 from both ing signaling events activating transcription factors (TFs) targeting sexes. We also generated these profiles from homozygous Ret null diverse genes and pathways varying across development (7, 8). -

Downloaded Per Proteome Cohort Via the Web- Site Links of Table 1, Also Providing Information on the Deposited Spectral Datasets
www.nature.com/scientificreports OPEN Assessment of a complete and classifed platelet proteome from genome‑wide transcripts of human platelets and megakaryocytes covering platelet functions Jingnan Huang1,2*, Frauke Swieringa1,2,9, Fiorella A. Solari2,9, Isabella Provenzale1, Luigi Grassi3, Ilaria De Simone1, Constance C. F. M. J. Baaten1,4, Rachel Cavill5, Albert Sickmann2,6,7,9, Mattia Frontini3,8,9 & Johan W. M. Heemskerk1,9* Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome‑wide transcriptomes (57.8 k mRNAs). For 14.8 k protein‑coding transcripts, we assigned the proteins to 21 UniProt‑based classes, based on their preferential intracellular localization and presumed function. This classifed transcriptome‑proteome profle of platelets revealed: (i) Absence of 37.2 k genome‑ wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein‑coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43–0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identifed proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma‑derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identifed proteome of nuclear‑related, membrane and signaling proteins, as well proteins with low‑level transcripts. -

The Expression Profiles of Mrnas and Lncrnas in Buffalo Muscle Stem Cells Driving Myogenic Differentiation
ORIGINAL RESEARCH published: 07 July 2021 doi: 10.3389/fgene.2021.643497 The Expression Profiles of mRNAs and lncRNAs in Buffalo Muscle Stem Cells Driving Myogenic Differentiation Ruimen Zhang 1†, Jinling Wang 1†, Zhengzhong Xiao 2†, Chaoxia Zou 1, Qiang An 1, Hui Li 1, Xiaoqing Zhou 2, Zhuyue Wu 2, Deshun Shi 1, Yanfei Deng 1*, Sufang Yang 1,3* and Yingming Wei 1* 1 State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Animal Reproduction Institute, Guangxi University, Nanning, China, 2 The Animal Husbandry Research Institute of Guangxi Autonomous, Nanning, China, 3 International Zhuang Medical Hospital Affiliated to Guangxi University Chinese Medicine, Nanning, China Edited by: Guohua Hua, Huazhong Agricultural Buffalo breeding has become an important branch of the beef cattle industry. Hence, it University, China is of great significance to study buffalo meat production and meat quality. However, the Reviewed by: expression profiles of mRNA and long non-coding RNAs (lncRNA) molecules in muscle Ikhide G. Imumorin, stem cells (MuSCs) development in buffalo have not been explored fully. We, therefore, Georgia Institute of Technology, United States performed mRNA and lncRNA expression profiling analysis during the proliferation Bo Wang, and differentiation phases of MuSCs in buffalo. The results showed that there were China Agricultural University, China 4,820 differentially expressed genes as well as 12,227 mRNAs and 1,352 lncRNAs. *Correspondence: Yanfei Deng These genes were shown to be enriched in essential biological processes such as [email protected] cell cycle, p53 signaling pathway, RNA transport and calcium signaling pathway. We Sufang Yang also identified a number of functionally important genes, such as MCMC4, SERDINE1, [email protected] Yingming Wei ISLR, LOC102394806, and LOC102403551, and found that interference with MYLPF [email protected] expression significantly inhibited the differentiation of MuSCs. -

Skeletal Muscle Transcriptome in Healthy Aging
ARTICLE https://doi.org/10.1038/s41467-021-22168-2 OPEN Skeletal muscle transcriptome in healthy aging Robert A. Tumasian III 1, Abhinav Harish1, Gautam Kundu1, Jen-Hao Yang1, Ceereena Ubaida-Mohien1, Marta Gonzalez-Freire1, Mary Kaileh1, Linda M. Zukley1, Chee W. Chia1, Alexey Lyashkov1, William H. Wood III1, ✉ Yulan Piao1, Christopher Coletta1, Jun Ding1, Myriam Gorospe1, Ranjan Sen1, Supriyo De1 & Luigi Ferrucci 1 Age-associated changes in gene expression in skeletal muscle of healthy individuals reflect accumulation of damage and compensatory adaptations to preserve tissue integrity. To characterize these changes, RNA was extracted and sequenced from muscle biopsies col- 1234567890():,; lected from 53 healthy individuals (22–83 years old) of the GESTALT study of the National Institute on Aging–NIH. Expression levels of 57,205 protein-coding and non-coding RNAs were studied as a function of aging by linear and negative binomial regression models. From both models, 1134 RNAs changed significantly with age. The most differentially abundant mRNAs encoded proteins implicated in several age-related processes, including cellular senescence, insulin signaling, and myogenesis. Specific mRNA isoforms that changed sig- nificantly with age in skeletal muscle were enriched for proteins involved in oxidative phosphorylation and adipogenesis. Our study establishes a detailed framework of the global transcriptome and mRNA isoforms that govern muscle damage and homeostasis with age. ✉ 1 National Institute on Aging–Intramural Research Program, National -

Rabbit Anti-MYLPF/FITC Conjugated Antibody
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-MYLPF/FITC Conjugated antibody SL5159R-FITC Product Name: Anti-MYLPF/FITC Chinese Name: FITC标记的快速骨骼肌肌球蛋白轻链2抗体 2410014J02Rik; DKFZp779C0757; DTNB; Fast skeletal myosin light chain 2; G2; HUMMLC2B; MGC13450; MLC 2; MRLC2; MYL11; MLC2B; MLC2F; Alias: MLRS_HUMAN; MYLPF; Myosin light polypeptide 2 alkali; Myosin regulatory light chain 2, skeletal muscle isoform. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Pig,Cow,Horse,Rabbit,Sheep, IF=1:50-200 Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 19kDa Form: Lyophilized or Liquid Concentration: 1mg/ml KLH conjugated synthetic peptide derived from human Fast skeletal myosin light chain immunogen: 2 Lsotype: IgGwww.sunlongbiotech.com Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. background: Myosin is a highly conserved, ubiquitously expressed protein that interacts with Actin to generate the force for cellular movements. Conventional Myosins are hexameric Product Detail: proteins consisting of two heavy chain subunits, a pair of non-phosphorylatable light chain subunits and a pair of phosphorylatable light chain subunits. -

Milger Et Al. Pulmonary CCR2+CD4+ T Cells Are Immune Regulatory And
Milger et al. Pulmonary CCR2+CD4+ T cells are immune regulatory and attenuate lung fibrosis development Supplemental Table S1 List of significantly regulated mRNAs between CCR2+ and CCR2- CD4+ Tcells on Affymetrix Mouse Gene ST 1.0 array. Genewise testing for differential expression by limma t-test and Benjamini-Hochberg multiple testing correction (FDR < 10%). Ratio, significant FDR<10% Probeset Gene symbol or ID Gene Title Entrez rawp BH (1680) 10590631 Ccr2 chemokine (C-C motif) receptor 2 12772 3.27E-09 1.33E-05 9.72 10547590 Klrg1 killer cell lectin-like receptor subfamily G, member 1 50928 1.17E-07 1.23E-04 6.57 10450154 H2-Aa histocompatibility 2, class II antigen A, alpha 14960 2.83E-07 1.71E-04 6.31 10590628 Ccr3 chemokine (C-C motif) receptor 3 12771 1.46E-07 1.30E-04 5.93 10519983 Fgl2 fibrinogen-like protein 2 14190 9.18E-08 1.09E-04 5.49 10349603 Il10 interleukin 10 16153 7.67E-06 1.29E-03 5.28 10590635 Ccr5 chemokine (C-C motif) receptor 5 /// chemokine (C-C motif) receptor 2 12774 5.64E-08 7.64E-05 5.02 10598013 Ccr5 chemokine (C-C motif) receptor 5 /// chemokine (C-C motif) receptor 2 12774 5.64E-08 7.64E-05 5.02 10475517 AA467197 expressed sequence AA467197 /// microRNA 147 433470 7.32E-04 2.68E-02 4.96 10503098 Lyn Yamaguchi sarcoma viral (v-yes-1) oncogene homolog 17096 3.98E-08 6.65E-05 4.89 10345791 Il1rl1 interleukin 1 receptor-like 1 17082 6.25E-08 8.08E-05 4.78 10580077 Rln3 relaxin 3 212108 7.77E-04 2.81E-02 4.77 10523156 Cxcl2 chemokine (C-X-C motif) ligand 2 20310 6.00E-04 2.35E-02 4.55 10456005 Cd74 CD74 antigen -

Transcriptomic and Proteomic Analysis of Hemidactylus Frenatus During Initial Stages of Tail Regeneration Sai Pawan Nagumantri, Sarena Banu & Mohammed M
www.nature.com/scientificreports OPEN Transcriptomic and proteomic analysis of Hemidactylus frenatus during initial stages of tail regeneration Sai Pawan Nagumantri, Sarena Banu & Mohammed M. Idris* Epimorphic regeneration of appendages is a complex and complete phenomenon found in selected animals. Hemidactylus frenatus, house gecko has the remarkable ability to regenerate the tail tissue upon autotomy involving epimorphic regeneration mechanism. This study has identifed and evaluated the molecular changes at gene and protein level during the initial stages, i.e., during the wound healing and repair mechanism initiation stage of tail regeneration. Based on next generation transcriptomics and De novo analysis the transcriptome library of the gecko tail tissue was generated. A total of 254 genes and 128 proteins were found to be associated with the regeneration of gecko tail tissue upon amputation at 1, 2 and 5-day post amputation (dpa) against control, 0-dpa through diferential transcriptomic and proteomic analysis. To authenticate the expression analysis, 50 genes were further validated involving RTPCR. 327 genes/proteins identifed and mapped from the study showed association for Protein kinase A signaling, Telomerase BAG2 signaling, paxillin signaling, VEGF signaling network pathways based on network pathway analysis. This study empanelled list of transcriptome, proteome and the list of genes/proteins associated with the tail regeneration. Regeneration is a sequential process specifcally controlled by cellular mechanisms to repair or replace tissue or organ from an injury. Te mass of undiferentiated cells (blastema) surrounding the injured tissue results in the formation of fully functional replica or imperfect replica which enacts the phenomenon of epimorphic regeneration1,2. -

Gene Expression Profile in Similar Tissues Using Transcriptome Sequencing Data of Whole-Body Horse Skeletal Muscle
G C A T T A C G G C A T genes Article Gene Expression Profile in Similar Tissues Using Transcriptome Sequencing Data of Whole-Body Horse Skeletal Muscle Ho-Yeon Lee 1,2, Jae-Yoon Kim 1,2, Kyoung Hyoun Kim 1,2 , Seongmun Jeong 1 , Youngbum Cho 1,2 and Namshin Kim 1,2,* 1 Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 34141, Korea; [email protected] (H.-Y.L.); [email protected] (J.-Y.K.); [email protected] (K.H.K.); [email protected] (S.J.); [email protected] (Y.C.) 2 Department of Bioinformatics, KRIBB School of Bioscience, University of Science and Technology (UST), Daejeon 34141, Korea * Correspondence: [email protected]; Tel.: +82-42-879-8162 Received: 8 October 2020; Accepted: 14 November 2020; Published: 17 November 2020 Abstract: Horses have been studied for exercise function rather than food production, unlike most livestock. Therefore, the role and characteristics of tissue landscapes are critically understudied, except for certain muscles used in exercise-related studies. In the present study, we compared RNA-Seq data from 18 Jeju horse skeletal muscles to identify differentially expressed genes (DEGs) between tissues that have similar functions and to characterize these differences. We identified DEGs between different muscles using pairwise differential expression (DE) analyses of tissue transcriptome expression data and classified the samples using the expression values of those genes. Each tissue was largely classified into two groups and their subgroups by k-means clustering, and the DEGs identified in comparison between each group were analyzed by functional/pathway level using gene set enrichment analysis and gene level, confirming the expression of significant genes. -
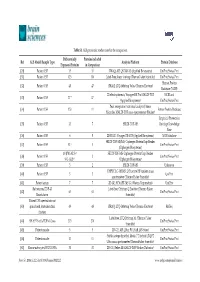
Table S1. ALS Proteomic Studies Used in the Comparison. Ref ALS Model/Sample Type Differentially Expressed Proteins Proteins
Table S1. ALS proteomic studies used in the comparison. Differentially Proteins included Ref ALS Model/Sample Type Analysis Platform Protein Database Expressed Proteins in Comparison [20] Patient CSF 35 35 iTRAQ; API QSTAR XL (Applied Biosystems) UniProt/Swiss-Prot [31] Patient CSF 123 110 Label-Free; linear ion trap (ThermoFisher Scientific) UniProt/Swiss-Prot Human Protein [32] Patient CSF 48 47 iTRAQ; LTQ-Orbitrap Velos (Thermo Electron) Database (NCBI) 2D electrophoresis; Voyager-DE Pro MALDI-TOF NCBI and [33] Patient CSF 17 * 17 (Applied Biosystems) UniProt/Swiss-Prot Peak recognition (statistical analysis); linear [34] Patient CSF 153 11 Entrez Protein Database Microflex MALDI-TOF mass spectrometer (Bruker) Empirical Proteomics [35] Patient CSF 33 7 SELDI-TOF-MS Ontology Knowledge Base [36] Patient CSF 6 5 2D-DIGE; Voyager DE-STR (Applied Biosystems) NCBI database SELDI-TOF-MS/MS; Ciphergen ProteinChip Reader [37] Patient CSF 52 † 3 UniProt/Swiss-Prot (Ciphergen Biosystems) 10 (PM-ALS) † SELDI-TOF-MS; Ciphergen ProteinChip Reader [38] Patient CSF 2 UniProt/Swiss-Prot 9 (L-ALS) † (Ciphergen Biosystems) [39] Patient CSF 3 2 SELDI-TOF-MS Unknown UHPLC LC-MS/MS; Q Exactive HF tandem mass [40] Patient CSF 3 3 UniProt spectrometer (Thermo Fisher Scientific) [41] Patient serum 7 7 2D-GE; SYNAPT-MS G1 (Waters Corporation) UniProt Rat neurons/TDP-43 Label-free; Orbitrap Q Exactive (Thermo Fisher [42] 63 63 UniProt/Swiss-Prot Knockdown Scientific) Patient CSF injected into rat [43] spinal cord /mitochondrial 49 48 iTRAQ; LTQ-Orbitrap Velos (Thermo Electron) RefSeq fraction Label-free; LTQ Orbitrap XL (Thermo Fisher [44] SH-SY5Y cells/TDP-43 Loss 273 270 UniProt/Swiss-Prot Scientific) [45] Patient muscle 5 5 2D-GE; API QStar PULSAR (AB-Sciex) UniProt/Swiss-Prot Stable-isotope dimethyl labels; 7 T hybrid LTQ FT [46] Patient muscle 11 11 UniProt/Swiss-Prot Ultra mass spectrometer (ThermoFisher Scientific) [47] Mouse astrocytes/SOD1 G93A 31 31 2D-GE; Reflex III MALDI-TOF (Bruker Daltonics) UniProt/Swiss-Prot Brain Sci.