Figure S1 Effect of Enzalutamide Treatment to Lncap and C4-2B Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -

Investigation of the Fatty Acid Transporter-Encoding Genes
www.nature.com/scientificreports OPEN Investigation of the fatty acid transporter-encoding genes SLC27A3 and SLC27A4 in autism Received: 13 February 2015 1 1 1 1 Accepted: 12 October 2015 Motoko Maekawa , Yoshimi Iwayama , Tetsuo Ohnishi , Manabu Toyoshima , 1 1 1 1 2,3 Published: 09 November 2015 Chie Shimamoto , Yasuko Hisano , Tomoko Toyota , Shabeesh Balan , Hideo Matsuzaki , Yasuhide Iwata3, Shu Takagai3, Kohei Yamada3, Motonori Ota4, Satoshi Fukuchi5, Yohei Okada6,7, Wado Akamatsu6,8, Masatsugu Tsujii3,9, Nobuhiko Kojima10, Yuji Owada11, Hideyuki Okano6, Norio Mori3 & Takeo Yoshikawa1 The solute carrier 27A (SLC27A) gene family encodes fatty acid transport proteins (FATPs) and includes 6 members. During fetal and postnatal periods of development, the growing brain requires a reliable supply of fatty acids. Because autism spectrum disorders (ASD) are now recognized as disorders caused by impaired early brain development, it is possible that functional abnormalities of SLC27A genes may contribute to the pathogenesis of ASD. Here, we confirmed the expression of SLC27A3 and SLC27A4 in human neural stem cells derived from human induced pluripotent stem cells, which suggested their involvement in the developmental stage of the central nervous system. Additionally, we resequenced the SLC27A3 and SLC27A4 genes using 267 ASD patient and 1140 control samples and detected 47 (44 novel and 29 nonsynonymous) and 30 (17 novel and 14 nonsynonymous) variants for the SLC27A3 and SLC27A4, respectively, revealing that they are highly polymorphic with multiple rare variants. The SLC27A4 Ser209 allele was more frequently represented in ASD samples. Furthermore, we showed that a SLC27A4 Ser209 mutant resulted in significantly higher fluorescently-labeled fatty acid uptake into bEnd3 cells, a mouse brain capillary-derived endothelial cell line, compared with SLC27A4 Gly209, suggesting that the functional change may contribute to ASD pathophysiology. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Phosphate Transporters of the SLC20 and SLC34 Families
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2013 Phosphate transporters of the SLC20 and SLC34 families Forster, Ian C ; Hernando, Nati ; Biber, Jürg ; Murer, Heini Abstract: Transport of inorganic phosphate (Pi) across the plasma membrane is essential for normal cellular function. Members of two families of SLC proteins (SLC20 and SLC34) act as Na(+)-dependent, secondary-active cotransporters to transport Pi across cell membranes. The SLC34 proteins are ex- pressed in specific organs important for Pi homeostasis: NaPi-IIa (SLC34A1) and NaPi-IIc (SLC34A3) fulfill essential roles in Pi reabsorption in the kidney proximal tubule and NaPi-IIb (SLC34A2) medi- ates Pi absorption in the gut. The SLC20 proteins, PiT-1 (SLC20A1), PiT-2 (SLC20A2) are expressed ubiquitously in all tissues and although generally considered as ”housekeeping” transport proteins, the discovery of tissue-specific activity, regulatory pathways and gene-related pathophysiologies, is redefining their importance. This review summarizes our current knowledge of SLC20 and SLC34 proteins in terms of their basic molecular characteristics, physiological roles, known pathophysiology and pharmacology. DOI: https://doi.org/10.1016/j.mam.2012.07.007 Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-79439 Journal Article Accepted Version Originally published at: Forster, Ian C; Hernando, Nati; Biber, Jürg; Murer, Heini (2013). Phosphate transporters of the SLC20 and SLC34 families. Molecular Aspects of Medicine, 34(2-3):386-395. DOI: https://doi.org/10.1016/j.mam.2012.07.007 1 Mini-reviews Series-Molecular Aspects of Medicine (JMAM) Phosphate transporters of the SLC20 and SLC34 families Ian C. -

(SLC22A18) (NM 183233) Human Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP304889 Solute carrier family 22 member 18 (SLC22A18) (NM_183233) Human Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Recombinant protein of human solute carrier family 22, member 18 (SLC22A18), transcript variant 2 Species: Human Expression Host: HEK293T Tag: C-Myc/DDK Predicted MW: 44.7 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol Preparation: Recombinant protein was captured through anti-DDK affinity column followed by conventional chromatography steps. Storage: Store at -80°C. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_899056 Locus ID: 5002 UniProt ID: Q96BI1 RefSeq Size: 1563 Cytogenetics: 11p15.4 RefSeq ORF: 1272 Synonyms: BWR1A; BWSCR1A; HET; IMPT1; ITM; ORCTL2; p45-BWR1A; SLC22A1L; TSSC5 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 Solute carrier family 22 member 18 (SLC22A18) (NM_183233) Human Recombinant Protein – TP304889 Summary: This gene is one of several tumor-suppressing subtransferable fragments located in the imprinted gene domain of 11p15.5, an important tumor-suppressor gene region. Alterations in this region have been associated with the Beckwith-Wiedemann syndrome, Wilms tumor, rhabdomyosarcoma, adrenocortical carcinoma, and lung, ovarian, and breast cancer. -

The Role of the Mtor Pathway in Developmental Reprogramming Of
THE ROLE OF THE MTOR PATHWAY IN DEVELOPMENTAL REPROGRAMMING OF HEPATIC LIPID METABOLISM AND THE HEPATIC TRANSCRIPTOME AFTER EXPOSURE TO 2,2',4,4'- TETRABROMODIPHENYL ETHER (BDE-47) An Honors Thesis Presented By JOSEPH PAUL MCGAUNN Approved as to style and content by: ________________________________________________________** Alexander Suvorov 05/18/20 10:40 ** Chair ________________________________________________________** Laura V Danai 05/18/20 10:51 ** Committee Member ________________________________________________________** Scott C Garman 05/18/20 10:57 ** Honors Program Director ABSTRACT An emerging hypothesis links the epidemic of metabolic diseases, such as non-alcoholic fatty liver disease (NAFLD) and diabetes with chemical exposures during development. Evidence from our lab and others suggests that developmental exposure to environmentally prevalent flame-retardant BDE47 may permanently reprogram hepatic lipid metabolism, resulting in an NAFLD-like phenotype. Additionally, we have demonstrated that BDE-47 alters the activity of both mTOR complexes (mTORC1 and 2) in hepatocytes. The mTOR pathway integrates environmental information from different signaling pathways, and regulates key cellular functions such as lipid metabolism, innate immunity, and ribosome biogenesis. Thus, we hypothesized that the developmental effects of BDE-47 on liver lipid metabolism are mTOR-dependent. To assess this, we generated mice with liver-specific deletions of mTORC1 or mTORC2 and exposed these mice and their respective controls perinatally to -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -
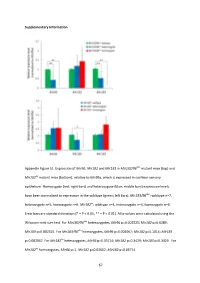
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

KCC4/SLC12A7 Polyclonal Antibody Catalog Number:26590-1-AP
For Research Use Only KCC4/SLC12A7 Polyclonal antibody www.ptglab.com Catalog Number:26590-1-AP Catalog Number: GenBank Accession Number: Purification Method: Basic Information 26590-1-AP BC098390 Antigen affinity purification Size: GeneID (NCBI): Recommended Dilutions: 150ul , Concentration: 900 μg/ml by 10723 WB 1:2000-1:10000 Nanodrop; Full Name: Source: solute carrier family 12 Rabbit (potassium/chloride transporters), Isotype: member 7 IgG Immunogen Catalog Number: AG24954 Applications Tested Applications: Positive Controls: WB, ELISA WB : BGC-823 cells, NCI-H1299 cells, Raji cells, SH- Species Specificity: SY5Y cells human KCC4, also named as Solute carrier family 12 member 7(SLC12A7), is a 1083 amino-acid protein, which belongs to Background Information the SLC12A transporter family. As a multi-pass membrane protein, the molecular weight of KCC4 is 119 kDa. KCC4 mediates electroneutral potassium-chloride cotransport when activated by cell swelling. Moreover, KCC4 may mediate K+ uptake into Deiters' cells in the cochlea and contribute to K+ recycling in the inner ear. KCC4 is important for the survival of cochlear outer and inner hair cells and the maintenance of the organ of Corti(Uniprot, GeneID:10723). Storage: Storage Store at -20°C. Stable for one year after shipment. Storage Buffer: PBS with 0.02% sodium azide and 50% glycerol pH 7.3. Aliquoting is unnecessary for -20ºC storage For technical support and original validation data for this product please contact: This product is exclusively available under Proteintech T: 1 (888) 4PTGLAB (1-888-478-4522) (toll free E: [email protected] Group brand and is not available to purchase from any in USA), or 1(312) 455-8498 (outside USA) W: ptglab.com other manufacturer.