Semantic Data-Analysis and Automated Decision Support Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Package 'Brotli'
Package ‘brotli’ May 13, 2018 Type Package Title A Compression Format Optimized for the Web Version 1.2 Description A lossless compressed data format that uses a combination of the LZ77 algorithm and Huffman coding. Brotli is similar in speed to deflate (gzip) but offers more dense compression. License MIT + file LICENSE URL https://tools.ietf.org/html/rfc7932 (spec) https://github.com/google/brotli#readme (upstream) http://github.com/jeroen/brotli#read (devel) BugReports http://github.com/jeroen/brotli/issues VignetteBuilder knitr, R.rsp Suggests spelling, knitr, R.rsp, microbenchmark, rmarkdown, ggplot2 RoxygenNote 6.0.1 Language en-US NeedsCompilation yes Author Jeroen Ooms [aut, cre] (<https://orcid.org/0000-0002-4035-0289>), Google, Inc [aut, cph] (Brotli C++ library) Maintainer Jeroen Ooms <[email protected]> Repository CRAN Date/Publication 2018-05-13 20:31:43 UTC R topics documented: brotli . .2 Index 4 1 2 brotli brotli Brotli Compression Description Brotli is a compression algorithm optimized for the web, in particular small text documents. Usage brotli_compress(buf, quality = 11, window = 22) brotli_decompress(buf) Arguments buf raw vector with data to compress/decompress quality value between 0 and 11 window log of window size Details Brotli decompression is at least as fast as for gzip while significantly improving the compression ratio. The price we pay is that compression is much slower than gzip. Brotli is therefore most effective for serving static content such as fonts and html pages. For binary (non-text) data, the compression ratio of Brotli usually does not beat bz2 or xz (lzma), however decompression for these algorithms is too slow for browsers in e.g. -
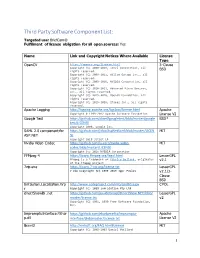
Third Party Software Component List: Targeted Use: Briefcam® Fulfillment of License Obligation for All Open Sources: Yes
Third Party Software Component List: Targeted use: BriefCam® Fulfillment of license obligation for all open sources: Yes Name Link and Copyright Notices Where Available License Type OpenCV https://opencv.org/license.html 3-Clause Copyright (C) 2000-2019, Intel Corporation, all BSD rights reserved. Copyright (C) 2009-2011, Willow Garage Inc., all rights reserved. Copyright (C) 2009-2016, NVIDIA Corporation, all rights reserved. Copyright (C) 2010-2013, Advanced Micro Devices, Inc., all rights reserved. Copyright (C) 2015-2016, OpenCV Foundation, all rights reserved. Copyright (C) 2015-2016, Itseez Inc., all rights reserved. Apache Logging http://logging.apache.org/log4cxx/license.html Apache Copyright © 1999-2012 Apache Software Foundation License V2 Google Test https://github.com/abseil/googletest/blob/master/google BSD* test/LICENSE Copyright 2008, Google Inc. SAML 2.0 component for https://github.com/jitbit/AspNetSaml/blob/master/LICEN MIT ASP.NET SE Copyright 2018 Jitbit LP Nvidia Video Codec https://github.com/lu-zero/nvidia-video- MIT codec/blob/master/LICENSE Copyright (c) 2016 NVIDIA Corporation FFMpeg 4 https://www.ffmpeg.org/legal.html LesserGPL FFmpeg is a trademark of Fabrice Bellard, originator v2.1 of the FFmpeg project 7zip.exe https://www.7-zip.org/license.txt LesserGPL 7-Zip Copyright (C) 1999-2019 Igor Pavlov v2.1/3- Clause BSD Infralution.Localization.Wp http://www.codeproject.com/info/cpol10.aspx CPOL f Copyright (C) 2018 Infralution Pty Ltd directShowlib .net https://github.com/pauldotknopf/DirectShow.NET/blob/ LesserGPL -

ROOT I/O Compression Improvements for HEP Analysis
EPJ Web of Conferences 245, 02017 (2020) https://doi.org/10.1051/epjconf/202024502017 CHEP 2019 ROOT I/O compression improvements for HEP analysis Oksana Shadura1;∗ Brian Paul Bockelman2;∗∗ Philippe Canal3;∗∗∗ Danilo Piparo4;∗∗∗∗ and Zhe Zhang1;y 1University of Nebraska-Lincoln, 1400 R St, Lincoln, NE 68588, United States 2Morgridge Institute for Research, 330 N Orchard St, Madison, WI 53715, United States 3Fermilab, Kirk Road and Pine St, Batavia, IL 60510, United States 4CERN, Meyrin 1211, Geneve, Switzerland Abstract. We overview recent changes in the ROOT I/O system, enhancing it by improving its performance and interaction with other data analysis ecosys- tems. Both the newly introduced compression algorithms, the much faster bulk I/O data path, and a few additional techniques have the potential to significantly improve experiment’s software performance. The need for efficient lossless data compression has grown significantly as the amount of HEP data collected, transmitted, and stored has dramatically in- creased over the last couple of years. While compression reduces storage space and, potentially, I/O bandwidth usage, it should not be applied blindly, because there are significant trade-offs between the increased CPU cost for reading and writing files and the reduces storage space. 1 Introduction In the past years, Large Hadron Collider (LHC) experiments are managing about an exabyte of storage for analysis purposes, approximately half of which is stored on tape storages for archival purposes, and half is used for traditional disk storage. Meanwhile for High Lumi- nosity Large Hadron Collider (HL-LHC) storage requirements per year are expected to be increased by a factor of 10 [1]. -
![Arxiv:2004.10531V1 [Cs.OH] 8 Apr 2020](https://docslib.b-cdn.net/cover/5419/arxiv-2004-10531v1-cs-oh-8-apr-2020-215419.webp)
Arxiv:2004.10531V1 [Cs.OH] 8 Apr 2020
ROOT I/O compression improvements for HEP analysis Oksana Shadura1;∗ Brian Paul Bockelman2;∗∗ Philippe Canal3;∗∗∗ Danilo Piparo4;∗∗∗∗ and Zhe Zhang1;y 1University of Nebraska-Lincoln, 1400 R St, Lincoln, NE 68588, United States 2Morgridge Institute for Research, 330 N Orchard St, Madison, WI 53715, United States 3Fermilab, Kirk Road and Pine St, Batavia, IL 60510, United States 4CERN, Meyrin 1211, Geneve, Switzerland Abstract. We overview recent changes in the ROOT I/O system, increasing per- formance and enhancing it and improving its interaction with other data analy- sis ecosystems. Both the newly introduced compression algorithms, the much faster bulk I/O data path, and a few additional techniques have the potential to significantly to improve experiment’s software performance. The need for efficient lossless data compression has grown significantly as the amount of HEP data collected, transmitted, and stored has dramatically in- creased during the LHC era. While compression reduces storage space and, potentially, I/O bandwidth usage, it should not be applied blindly: there are sig- nificant trade-offs between the increased CPU cost for reading and writing files and the reduce storage space. 1 Introduction In the past years LHC experiments are commissioned and now manages about an exabyte of storage for analysis purposes, approximately half of which is used for archival purposes, and half is used for traditional disk storage. Meanwhile for HL-LHC storage requirements per year are expected to be increased by factor 10 [1]. arXiv:2004.10531v1 [cs.OH] 8 Apr 2020 Looking at these predictions, we would like to state that storage will remain one of the major cost drivers and at the same time the bottlenecks for HEP computing. -

A Novel Coding Architecture for Multi-Line Lidar Point Clouds
This article has been accepted for inclusion in a future issue of this journal. Content is final as presented, with the exception of pagination. IEEE TRANSACTIONS ON INTELLIGENT TRANSPORTATION SYSTEMS 1 A Novel Coding Architecture for Multi-Line LiDAR Point Clouds Based on Clustering and Convolutional LSTM Network Xuebin Sun , Sukai Wang , Graduate Student Member, IEEE, and Ming Liu , Senior Member, IEEE Abstract— Light detection and ranging (LiDAR) plays an preservation of historical relics, 3D sensing for smart city, indispensable role in autonomous driving technologies, such as well as autonomous driving. Especially for autonomous as localization, map building, navigation and object avoidance. driving systems, LiDAR sensors play an indispensable role However, due to the vast amount of data, transmission and storage could become an important bottleneck. In this article, in a large number of key techniques, such as simultaneous we propose a novel compression architecture for multi-line localization and mapping (SLAM) [1], path planning [2], LiDAR point cloud sequences based on clustering and convolu- obstacle avoidance [3], and navigation. A point cloud consists tional long short-term memory (LSTM) networks. LiDAR point of a set of individual 3D points, in accordance with one or clouds are structured, which provides an opportunity to convert more attributes (color, reflectance, surface normal, etc). For the 3D data to 2D array, represented as range images. Thus, we cast the 3D point clouds compression as a range image instance, the Velodyne HDL-64E LiDAR sensor generates a sequence compression problem. Inspired by the high efficiency point cloud of up to 2.2 billion points per second, with a video coding (HEVC) algorithm, we design a novel compression range of up to 120 m. -

I Introduction
PPM Performance with BWT Complexity: A New Metho d for Lossless Data Compression Michelle E ros California Institute of Technology e [email protected] Abstract This work combines a new fast context-search algorithm with the lossless source co ding mo dels of PPM to achieve a lossless data compression algorithm with the linear context-search complexity and memory of BWT and Ziv-Lemp el co des and the compression p erformance of PPM-based algorithms. Both se- quential and nonsequential enco ding are considered. The prop osed algorithm yields an average rate of 2.27 bits per character bp c on the Calgary corpus, comparing favorably to the 2.33 and 2.34 bp c of PPM5 and PPM and the 2.43 bp c of BW94 but not matching the 2.12 bp c of PPMZ9, which, at the time of this publication, gives the greatest compression of all algorithms rep orted on the Calgary corpus results page. The prop osed algorithm gives an average rate of 2.14 bp c on the Canterbury corpus. The Canterbury corpus web page gives average rates of 1.99 bp c for PPMZ9, 2.11 bp c for PPM5, 2.15 bp c for PPM7, and 2.23 bp c for BZIP2 a BWT-based co de on the same data set. I Intro duction The Burrows Wheeler Transform BWT [1] is a reversible sequence transformation that is b ecoming increasingly p opular for lossless data compression. The BWT rear- ranges the symb ols of a data sequence in order to group together all symb ols that share the same unb ounded history or \context." Intuitively, this op eration is achieved by forming a table in which each row is a distinct cyclic shift of the original data string. -

Dc5m United States Software in English Created at 2016-12-25 16:00
Announcement DC5m United States software in english 1 articles, created at 2016-12-25 16:00 articles set mostly positive rate 10.0 1 3.8 Google’s Brotli Compression Algorithm Lands to Windows Edge Microsoft has announced that its Edge browser has started using Brotli, the compression algorithm that Google open-sourced last year. 2016-12-25 05:00 1KB www.infoq.com Articles DC5m United States software in english 1 articles, created at 2016-12-25 16:00 1 /1 3.8 Google’s Brotli Compression Algorithm Lands to Windows Edge Microsoft has announced that its Edge browser has started using Brotli, the compression algorithm that Google open-sourced last year. Brotli is on by default in the latest Edge build and can be previewed via the Windows Insider Program. It will reach stable status early next year, says Microsoft. Microsoft touts a 20% higher compression ratios over comparable compression algorithms, which would benefit page load times without impacting client-side CPU costs. According to Google, Brotli uses a whole new data format , which makes it incompatible with Deflate but ensures higher compression ratios. In particular, Google says, Brotli is roughly as fast as zlib when decompressing and provides a better compression ratio than LZMA and bzip2 on the Canterbury Corpus. Brotli appears to be especially tuned for the web , that is for offline encoding and online decoding of Web assets, or Android APKs. Google claims a compression ratio improvement of 20–26% over its own Zopfli algorithm, which still provides the best compression ratio of any deflate algorithm. -

Nxadmin CLI Reference Guide Unity Iv Contents
HYPER-UNIFIED STORAGE nxadmin Command Line Interface Reference Guide NEXSAN | 325 E. Hillcrest Drive, Suite #150 | Thousand Oaks, CA 91360 USA Printed Thursday, July 26, 2018 | www.nexsan.com Copyright © 2010—2018 Nexsan Technologies, Inc. All rights reserved. Trademarks Nexsan® is a trademark or registered trademark of Nexsan Technologies, Inc. The Nexsan logo is a registered trademark of Nexsan Technologies, Inc. All other trademarks and registered trademarks are the property of their respective owners. Patents This product is protected by one or more of the following patents, and other pending patent applications worldwide: United States patents US8,191,841, US8,120,922; United Kingdom patents GB2466535B, GB2467622B, GB2467404B, GB2296798B, GB2297636B About this document Unauthorized use, duplication, or modification of this document in whole or in part without the written consent of Nexsan Corporation is strictly prohibited. Nexsan Technologies, Inc. reserves the right to make changes to this manual, as well as the equipment and software described in this manual, at any time without notice. This manual may contain links to web sites that were current at the time of publication, but have since been moved or become inactive. It may also contain links to sites owned and operated by third parties. Nexsan is not responsible for the content of any such third-party site. Contents Contents Contents iii Chapter 1: Accessing the nxadmin and nxcmd CLIs 15 Connecting to the Unity Storage System using SSH 15 Prerequisite 15 Connecting to the Unity -

Implementing Associative Coder of Buyanovsky (ACB)
Implementing Associative Coder of Buyanovsky (ACB) data compression by Sean Michael Lambert A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Computer Science Montana State University © Copyright by Sean Michael Lambert (1999) Abstract: In 1994 George Mechislavovich Buyanovsky published a basic description of a new data compression algorithm he called the “Associative Coder of Buyanovsky,” or ACB. The archive program using this idea, which he released in 1996 and updated in 1997, is still one of the best general compression utilities available. Despite this, the ACB algorithm is still barely understood by data compression experts, primarily because Buyanovsky never published a detailed description of it. ACB is a new idea in data compression, merging concepts from existing statistical and dictionary-based algorithms with entirely original ideas. This document presents several variations of the ACB algorithm and the details required to implement a basic version of ACB. IMPLEMENTING ASSOCIATIVE CODER OF BUYANOVSKY (ACB) DATA COMPRESSION by Sean Michael Lambert A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Computer Science MONTANA STATE UNIVERSITY-BOZEMAN Bozeman, Montana April 1999 © COPYRIGHT by Sean Michael Lambert 1999 All Rights Reserved ii APPROVAL of a thesis submitted by Sean Michael Lambert This thesis has been read by each member of the thesis committee and has been found to be satisfactory regarding content, English usage, format, citations, bibliographic style, and consistency, and is ready for submission to the College of Graduate Studies. Brendan Mumey U /l! ^ (Signature) Date Approved for the Department of Computer Science J. -

An Analysis of XML Compression Efficiency
An Analysis of XML Compression Efficiency Christopher J. Augeri1 Barry E. Mullins1 Leemon C. Baird III Dursun A. Bulutoglu2 Rusty O. Baldwin1 1Department of Electrical and Computer Engineering Department of Computer Science 2Department of Mathematics and Statistics United States Air Force Academy (USAFA) Air Force Institute of Technology (AFIT) USAFA, Colorado Springs, CO Wright Patterson Air Force Base, Dayton, OH {chris.augeri, barry.mullins}@afit.edu [email protected] {dursun.bulutoglu, rusty.baldwin}@afit.edu ABSTRACT We expand previous XML compression studies [9, 26, 34, 47] by XML simplifies data exchange among heterogeneous computers, proposing the XML file corpus and a combined efficiency metric. but it is notoriously verbose and has spawned the development of The corpus was assembled using guidelines given by developers many XML-specific compressors and binary formats. We present of the Canterbury corpus [3], files often used to assess compressor an XML test corpus and a combined efficiency metric integrating performance. The efficiency metric combines execution speed compression ratio and execution speed. We use this corpus and and compression ratio, enabling simultaneous assessment of these linear regression to assess 14 general-purpose and XML-specific metrics, versus prioritizing one metric over the other. We analyze compressors relative to the proposed metric. We also identify key collected metrics using linear regression models (ANOVA) versus factors when selecting a compressor. Our results show XMill or a simple comparison of means, e.g., X is 20% better than Y. WBXML may be useful in some instances, but a general-purpose compressor is often the best choice. 2. XML OVERVIEW XML has gained much acceptance since first proposed in 1998 by Categories and Subject Descriptors the World-Wide Web Consortium (W3C). -

Unfoldr Dstep
Asymmetric Numeral Systems Jeremy Gibbons WG2.11#19 Salem ANS 2 1. Coding Huffman coding (HC) • efficient; optimally effective for bit-sequence-per-symbol arithmetic coding (AC) • Shannon-optimal (fractional entropy); but computationally expensive asymmetric numeral systems (ANS) • efficiency of Huffman, effectiveness of arithmetic coding applications of streaming (another story) • ANS introduced by Jarek Duda (2006–2013). Now: Facebook (Zstandard), Apple (LZFSE), Google (Draco), Dropbox (DivANS). ANS 3 2. Intervals Pairs of rationals type Interval (Rational, Rational) = with operations unit (0, 1) = weight (l, r) x l (r l) x = + − ⇥ narrow i (p, q) (weight i p, weight i q) = scale (l, r) x (x l)/(r l) = − − widen i (p, q) (scale i p, scale i q) = so that narrow and unit form a monoid, and inverse relationships: weight i x i x unit 2 () 2 weight i x y scale i y x = () = narrow i j k widen i k j = () = ANS 4 3. Models Given counts :: [(Symbol, Integer)] get encodeSym :: Symbol Interval ! decodeSym :: Rational Symbol ! such that decodeSym x s x encodeSym s = () 2 1 1 1 1 Eg alphabet ‘a’, ‘b’, ‘c’ with counts 2, 3, 5 encoded as (0, /5), ( /5, /2), and ( /2, 1). { } ANS 5 4. Arithmetic coding encode1 :: [Symbol ] Rational ! encode1 pick foldl estep unit where = ◦ 1 estep :: Interval Symbol Interval 1 ! ! estep is narrow i (encodeSym s) 1 = decode1 :: Rational [Symbol ] ! decode1 unfoldr dstep where = 1 dstep :: Rational Maybe (Symbol, Rational) 1 ! dstep x let s decodeSym x in Just (s, scale (encodeSym s) x) 1 = = where pick :: Interval Rational satisfies pick i i. -

Improving Compression-Ratio in Backup
Institutionen för systemteknik Department of Electrical Engineering Examensarbete Improving compression ratio in backup Examensarbete utfört i Informationskodning/Bildkodning av Mattias Zeidlitz Författare Mattias Zeidlitz LITH-ISY-EX--12/4588--SE Linköping 2012 TEKNISKA HÖGSKOLAN LINKÖPINGS UNIVERSITET Department of Electrical Engineering Linköpings tekniska högskola Linköping University Institutionen för systemteknik S-581 83 Linköping, Sweden 581 83 Linköping Improving compression-ratio in backup ............................................................................ Examensarbete utfört i Informationskodning/Bildkodning vid Linköpings tekniska högskola av Mattias Zeidlitz ............................................................. LITH-ISY-EX--12/4588--SE Presentationsdatum Institution och avdelning 2012-06-13 Institutionen för systemteknik Publiceringsdatum (elektronisk version) Department of Electrical Engineering Datum då du ämnar publicera exjobbet Språk Typ av publikation ISBN (licentiatavhandling) Svenska Licentiatavhandling ISRN LITH-ISY-EX--12/4588--SE x Annat (ange nedan) x Examensarbete Serietitel (licentiatavhandling) C-uppsats D-uppsats Engelska Rapport Serienummer/ISSN (licentiatavhandling) Antal sidor Annat (ange nedan) 58 URL för elektronisk version http://www.ep.liu.se Publikationens titel Improving compression ratio in backup Författare Mattias Zeidlitz Sammanfattning Denna rapport beskriver ett examensarbete genomfört på Degoo Backup AB i Stockholm under våren 2012. Syftet var att designa en kompressionssvit