Allozyme Variation and Possible Phylogenetic Implications in Abies Cephalonica Loudon and Some Related Eastern Mediterranean Firs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Occurrence of Rhyndophorus Ferrugineus in Grecce and Cyprus
ENTOMOLOGIA HELLENICA 17 (2007-2008): 28-33 The scale insect Dynaspidiotus abietis (Schrank) on Abies cephalonica (Pinaceae) G. J. STATHAS Technological Educational Institute of Kalamata, School of Agricultural Technology Department of Crop Production, Laboratory of Agricultural Entomology and Zoology, 24100 Antikalamos, Greece, ([email protected]) ABSTRACT Data on phenology and morphology of the scale insect Dynaspidiotus abietis (Schrank) (Hemiptera: Diaspididae), found on fir trees Abies cephalonica (Pinaceae) on mount Taygetos (Peloponnesus - southern Greece), are presented. The species is biparental and oviparous. During this study (June 2004 – August 2006) D. abietis completed one generation per year. It overwintered as mated pre-ovipositing female adult. Ovipositions were recorded from May to July. The majority of the hatches of the crawlers were observed in June. Predated individuals of the scale which were found during the study period were attributed to the presence of the predator Chilocorus bipustulatus (L.) (Coleoptera: Coccinellidae). Introduction belonging to the family Coccidae, such as Physokermes hemicryphus (Dalm.), P. There are many species belonging to picae Sch., Eulecanium sericeum (Lind.) family Diaspididae that infest fir trees in and Nemolecanium graniformis (Wünn), Europe. Major species include: Chionaspis which were found on Abies cephalonica austriaca Lindinger, Diaspidiotus Loud. and A. borisii-regis Mattf., as well ostreaeformis (Curtis), Dynaspidiotus as Marchalina hellenica (Gennadius) abieticola (Koroneos), D. abietis (Margarodidae), are regarded as more (Schrank), Fiorinia japonica Luwana, important and have been studied mainly Lepidosaphes juniperi Lindinger, L. due to them excreting honeydews, on newsteadi (Šulc), Leucaspis lowi Colvée, which bees are fed (Santas 1983, Santas L. pini (Hartig), Parlatoria parlatoriae 1991, Stathas, 2001). (Šulc) and Unaspidiotus corticispini The scale insect Dynaspidiotus abietis (Lindinger) (Ben-Dov 2006). -

40YEARS 1980-2020 Check out Our Plugs
Brooks Tree Farm CELEBRATING 40YEARS 1980-2020 Check out our Plugs Norway Spruce is grown in much of the USA, it tolerates a wide range of soils and takes shearing well. Good cold climate Christmas tree, also used as understock for all Spruce grafting. Colorado Spruce has a wide variety of colors, some more blue than others. Native to the Rocky Mountains, but used all over as an ornamental species. Needles can be sharp to the touch. Ponderosa Pine grows very straight and tall and has long needles. Norway Spruce Colorado Spruce Ponderosa Pine The Willamette Valley seed source was nearly extinct, it tolerates wet soil. Mugo Mughus is a slow growing pine that reaches 8’. Ornamentally used in landscapes, embankments, rock walls, Mughus is very resistant to cold and drought. Salal is a low ornamental shrub. The leaves are often used by florists. Native to the west coast Salal is often found on Giantembankments Sequoia on is the a fast coast growing or on forest conifer floors. to 200’ from California, often grafted with weeping form. Also popular as large scale evergreen screen. Mugo Mughus Salal Giant Sequoia Root Dip Protect your investment by dipping roots in a super absorbent root protecting hydrogel prior Horta Sorb to planting. The gels attract moisture and hold it in the root zone keeping it accessible to the 1 Pound $25.00 roots during dry periods. The polymer gels can absorb hundreds of times their weight in 4 Pounds $80.00 water and release the moisture to the plant multiple times, providing water to the root zone 10 Pounds $150.00 and protecting the plants during roots establishment. -

ISTA List of Stabilized Plant Names 7Th Edition
ISTA List of Stabilized Plant Names th 7 Edition ISTA Nomenclature Committee Chair: Dr. M. Schori Published by All rights reserved. No part of this publication may be The Internation Seed Testing Association (ISTA) reproduced, stored in any retrieval system or transmitted Zürichstr. 50, CH-8303 Bassersdorf, Switzerland in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior ©2020 International Seed Testing Association (ISTA) permission in writing from ISTA. ISBN 978-3-906549-77-4 ISTA List of Stabilized Plant Names 1st Edition 1966 ISTA Nomenclature Committee Chair: Prof P. A. Linehan 2nd Edition 1983 ISTA Nomenclature Committee Chair: Dr. H. Pirson 3rd Edition 1988 ISTA Nomenclature Committee Chair: Dr. W. A. Brandenburg 4th Edition 2001 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 5th Edition 2007 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 6th Edition 2013 ISTA Nomenclature Committee Chair: Dr. J. H. Wiersema 7th Edition 2019 ISTA Nomenclature Committee Chair: Dr. M. Schori 2 7th Edition ISTA List of Stabilized Plant Names Content Preface .......................................................................................................................................................... 4 Acknowledgements ....................................................................................................................................... 6 Symbols and Abbreviations .......................................................................................................................... -
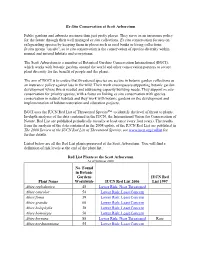
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Proceedings of the 9Th International Christmas Tree Research & Extension Conference
Proceedings of the 9th International Christmas Tree Research & Extension Conference September 13–18, 2009 _________________________________________________________________________________________________________ John Hart, Chal Landgren, and Gary Chastagner (eds.) Title Proceedings of the 9th International Christmas Tree Research & Extension Conference IUFRO Working Unit 2.02.09—Christmas Trees Corvallis, Oregon and Puyallup, Washington, September 13–18, 2009 Held by Oregon State University, Washington State University, and Pacific Northwest Christmas Tree Growers’ Association Editors John Hart Chal Landgren Gary Chastagner Compilation by Teresa Welch, Wild Iris Communications, Corvallis, OR Citation Hart, J., Landgren, C., and Chastagner, G. (eds.). 2010. Proceedings of the 9th International Christmas Tree Research and Extension Conference. Corvallis, OR and Puyallup, WA. Fair use This publication may be reproduced or used in its entirety for noncommercial purposes. Foreword The 9th International Christmas Tree Research and Extension Conference returned to the Pacific Northwest in 2009. OSU and WSU cohosted the conference, which was attended by 42 Christmas tree professionals representing most of the major production areas in North America and Europe. This conference was the most recent in the following sequence: Date Host Location Country October 1987 Washington State University Puyallup, Washington USA August 1989 Oregon State University Corvallis, Oregon USA October 1992 Oregon State University Silver Falls, Oregon USA September 1997 -

Rooting Nordmann Fir Cuttings for Christmas Trees?
Working Papers of the Finnish Forest Research Institute 114 Vegetative propagation of conifers for enhancing landscaping and tree breeding. Proceedings of the Nordic meeting held in September 10th-11th 2008 at Punkaharju, Finland http://www.metla.fi/julkaisut/workingpapers/2009/mwp114.htm Working Papers of the Finnish Forest Research Institute 114: 48–52 Rooting Nordmann fir cuttings for Christmas trees? Ulrik Bräuner Nielsen1, Hanne N. Rasmussen1 and Martin Jensen2 1 University of Copenhagen, Forest and Landscape, Denmark; [email protected] 2 University of Aarhus, Department of Horticulture, Aarslev, Denmark Recent succes with rooted cuttings in fir (Abies spp.) propagation stimulated this experiment with Abies nordmanniana, an important Christmas tree species in Denmark. Cuttings were taken in late summer from young and older trees, untreated or stumped, and from specified positions within the crown. Auxin was administered to some cuttings at varying concentrations. Rooting was monitored over a period of 6 months and attained 60-70% in the best shoot types. Auxin had no influence on rooting but concentration correlated positively with tissue decay in basal end of cuttings. Rooting of cuttings from the leader in untreated trees decreased markedly with age while cuttings from the branches decreased less dramatically. No basipetal trend of rooting capability of branches could be ascertained. Orthotropic shoots that regenerated on stumped trees rooted with varying success, the ones originating from the main stem, and preferably at a low position, performing best. Orthotropic growth was maintained in 35-45% of these shoots 8 months after rooting, while plagiotropic growth was prevalent in cuttings of other shoot types. -

Diversity in Needle Morphology and Genetic Markers in a Marginal Abies Cephalonica (Pinaceae) Population
Ann. For. Res. 58(2): 217-234, 2015 ANNALS OF FOREST RESEARCH DOI: 10.15287/afr.2015.410 www.afrjournal.org Diversity in needle morphology and genetic markers in a marginal Abies cephalonica (Pinaceae) population A.C. Papageorgiou, C. Kostoudi, I. Sorotos, G. Varsamis, G. Korakis, A.D. Drouzas Papageorgiou A.C., Kostoudi C., Sorotos I., Varsamis G., Korakis G., Drouzas A.D., 2015. Diversity in needle morphology and genetic markers in a marginal Abies cephalonica (Pinaceae) population. Ann. For. Res. 58(2): 217-234. Abstract. Differences in needle traits of coniferous tree species are consid- ered as the combined result of direct environmental pressure and specific genetic adaptations. In this study, diversity and differentiation within and among four Abies cephalonica subpopulations of a marginal population on Mt. Parnitha - Greece, were estimated using needle morphological traits and gene markers. We tested the connection of morphological variability patterns of light and shade needles with possible adaptation strategies and genetic diversity. Six morphological characteristics were used for the de- scription of both light and shade needles at 100 trees, describing needle size and shape, stomatal density and needle position on the twigs. Additionally, six RAPD and three ISSR markers were applied on DNA from the same trees. Light needles were significantly different than shade needles, in all traits measured, apparently following a different light harvesting strategy. All four subpopulations exhibited high genetic diversity and the differen- tiation among them was relatively low. Differences among populations in light needles seemed to depend on light exposure and aspect. In shade nee- dles, the four subpopulations seemed to deviate stronger from each other and express a rather geographic pattern, similarly to the genetic markers. -

Evaluating Mediterranean Firs for Use in Pennsylvania©
170 Combined Proceedings International Plant Propagators’ Society, Volume 58, 2008 Evaluating Mediterranean Firs for Use in Pennsylvania© Ricky M. Bates Department of Horticulture, The Pennsylvania State University, University Park, Pennsylvania 16802 U.S.A. Email: [email protected] David L. Sanford Department of Horticulture, The Pennsylvania State University, Berks Campus, Reading, Pennsylvania 19610 U.S.A. INTRODUCTION The true firs Abies( sp. Mill.) include over 40 tree species widely scattered throughout the northern hemisphere. Economically, firs remain underdeveloped in the U.S.A. as a landscape plant due to a general reputation for sensitivity to hot, dry, urban conditions and a lack of consistent and replicated evaluation across a broad range of environments and conditions. True firs are preferred as Christmas tree species by U.S.A. consumers due to their natural conical shape, pleasing aroma, stout branch structure, and generally excellent postharvest needle retention. Eastern U.S.A. Christmas tree growers have also been relying upon a very limited selection of fir species including Abies fraseri, A. balsamea, A. balsamea var. phanerolepis, and A. concolor. Unfortunately, all of the aforementioned native firs are extremely vulner- able to Phytophthora root rot and can be very site demanding (Frampton and Benson, 2004; Benson et al., 1998). Anecdotal evidence from garden and arboreta curators, horticulture researchers, and some non-replicated trials indicate that certain Abies species native to the Mediterranean region perform well under adverse conditions in the Mid-Atlantic and Northeast U.S.A. (Gutowski and Thomas, 1962). THE MEDITERRANEAN FIRS The Mediterranean firs comprise a group of approximately 10Abies species native to countries bordering, or in close proximity to, the Mediterranean Sea (Table 1). -

(Homoptera: Adelgidae) Galls on a Several Years Old Spruce (Picea Abies) Plantation
© Entomologica Fennica. 30 December 2014 Factors associated with the distribution and size of Adelges abietis (Homoptera: Adelgidae) galls on a several years old spruce (Picea abies) plantation Sebastian Pilichowski, Damian Pazio & Marian J. Giertych Pilichowski, S., Pazio, D. & Giertych, M. J. 2014: Factors associated with the distribution and size of Adelges abietis (Homoptera: Adelgidae) galls on a sev- eral years old spruce (Picea abies) plantation. Entomol. Fennica 25: 161169. The eastern spruce gall adelgid (Adelges abietis) is a homopteran insect related to aphids. It forms pineapple-like pseudocone galls on Norway spruce (Picea abies) shoots, which are often deformed by them. The factors responsible for gall distri- bution within a tree crown are not clear. We investigated the distribution of galls in crowns on a spruce plantation. We assumed that the number and size of galls is dependent on the size of the tree, position on the crown, the type of shoot and its placement relative to the compass points. We also counted the larval chambers and related their number with gall size. We demonstrated that the largest galls are in the upper part of the crown and on the lateral shoots, and the number of cham- bers is closely related to the size of the galls. The level of shoot embrace has a sig- nificant impact on shoot deformation. S. Pilichowski & D. Pazio, University of Zielona Góra, Faculty of Biological Sci- ences, Prof. Z. Szafrana 1; 65-516 Zielona Góra, Poland; E-mails: s.pilichowski @wnb.uz.zgora.pl, & [email protected] M. J. Giertych, University of Zielona Góra, Faculty of Biological Sciences, Prof. -

ABSTRACT JETTON, ROBERT MILLER. Biological Control, Host
ABSTRACT JETTON, ROBERT MILLER. Biological Control, Host Resistance, and Vegetative Propagation: Strategies and Tools for Management of the Invasive Hemlock Woolly Adelgid, Adelges tsugae Annand. (Under the direction of Dr. Fred P. Hain.) Biological control, host resistance, and vegetative propagation were evaluated as strategies and tools for management of the exotic pest hemlock woolly adelgid (HWA), Adelges tsugae Annand (Hemiptera: Adelgidae) in the southeastern U.S. The suitability of the balsam woolly adelgid (BWA), Adelges piceae Ratzeburg, as an alternate rearing host for Sasajiscymnus tsugae was compared to the primary prey HWA in a series of laboratory- based paired-choice and no choice (single-prey) experiments that tested adult feeding, oviposition, and long-term survival and immature development of the predator. Results indicated that S. tsugae will feed equally on eggs of both prey, will accept both prey for oviposition, and that the predator’s rate of immature development did not differ between the prey species, although fewer successfully completed egg to adult development on a diet of BWA compared to a diet of HWA. The long-term survival of predator adults was significantly influenced by both test prey type and the availability of a supplemental food source. The utility of confined releases for colonization of S. tsugae was evaluated in three field studies at forest and ornamental sites in western North Carolina. Predator reproduction, survival, and impact on HWA were investigated following the placement of fifteen adults (10♀:5♂) in mesh sleeves cages on adelgid infested hemlock branches for two or four weeks. In all three studies the predator reproduced inside sleeve cages and oviposition generally began within two or three weeks. -

The Evolution of Cavitation Resistance in Conifers Maximilian Larter
The evolution of cavitation resistance in conifers Maximilian Larter To cite this version: Maximilian Larter. The evolution of cavitation resistance in conifers. Bioclimatology. Univer- sit´ede Bordeaux, 2016. English. <NNT : 2016BORD0103>. <tel-01375936> HAL Id: tel-01375936 https://tel.archives-ouvertes.fr/tel-01375936 Submitted on 3 Oct 2016 HAL is a multi-disciplinary open access L'archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destin´eeau d´ep^otet `ala diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publi´esou non, lished or not. The documents may come from ´emanant des ´etablissements d'enseignement et de teaching and research institutions in France or recherche fran¸caisou ´etrangers,des laboratoires abroad, or from public or private research centers. publics ou priv´es. THESE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITE DE BORDEAUX Spécialité : Ecologie évolutive, fonctionnelle et des communautés Ecole doctorale: Sciences et Environnements Evolution de la résistance à la cavitation chez les conifères The evolution of cavitation resistance in conifers Maximilian LARTER Directeur : Sylvain DELZON (DR INRA) Co-Directeur : Jean-Christophe DOMEC (Professeur, BSA) Soutenue le 22/07/2016 Devant le jury composé de : Rapporteurs : Mme Amy ZANNE, Prof., George Washington University Mr Jordi MARTINEZ VILALTA, Prof., Universitat Autonoma de Barcelona Examinateurs : Mme Lisa WINGATE, CR INRA, UMR ISPA, Bordeaux Mr Jérôme CHAVE, DR CNRS, UMR EDB, Toulouse i ii Abstract Title: The evolution of cavitation resistance in conifers Abstract Forests worldwide are at increased risk of widespread mortality due to intense drought under current and future climate change. -

Euronatur Spezial 1 2014 Ancient Beech Forests in Albania And
Report of the Excursion to Ancient Beech Forests in Albania and Macedonia July 14-19, 2013 Hans D. Knapp, Christel Schroeder, Gabriel Schwaderer Albania, Shebenik-Jablanica National Park, Rajca, ancient beech forest with Allium ursinum © Hans D. Knapp EuroNatur Spezial 01/2014 Report of the Excursion to Ancient Beech Forests in Albania and Macedonia - 1 - 1. Reason and background EuroNatur is active on the Balkan Peninsula and especially in the Southwest Balkan since more than 20 years. Focus of the activities is the protection of the most valuable lakes and wetlands as well as the most important mountainous areas. Of particular natural value are the areas which were formerly part of the iron curtain. EuroNatur – together with many other organizations – has developed an initiative to protect these areas: the European Green Belt Initiative. Especially along the border of Albania, with Montenegro, with Kosovo and with Macedonia many landscapes deserve a protection status and therefore EuroNatur mainly with its partner organizations in Albania and Macedonia promotes the designation of National Parks and Nature Parks. Two National Parks and one Nature Park in Albania were already proclaimed based on these activities. In Macedonia two more National Parks were proposed but not designated yet. Protection and Preservation of Natural Environment in Albania (PPNEA) is a non-governmental environmental organization that operates in the entire territory of Albania. PPNEA is known to be the first environmental organization in Albania, as it was officially established on June 13th, 1991, with a special decree of the Albanian Academy of Sciences at the time. It emerged in a period of turmoil and socio-economic change in the country and the wider region - a time where environmental issues were looked down upon and given marginal priority in governing and policy issues.