82406414.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Shedding New Light on the Generation of the Visual Chromophore PERSPECTIVE Krzysztof Palczewskia,B,C,1 and Philip D
PERSPECTIVE Shedding new light on the generation of the visual chromophore PERSPECTIVE Krzysztof Palczewskia,b,c,1 and Philip D. Kiserb,d Edited by Jeremy Nathans, Johns Hopkins University School of Medicine, Baltimore, MD, and approved July 9, 2020 (received for review May 16, 2020) The visual phototransduction cascade begins with a cis–trans photoisomerization of a retinylidene chro- mophore associated with the visual pigments of rod and cone photoreceptors. Visual opsins release their all-trans-retinal chromophore following photoactivation, which necessitates the existence of pathways that produce 11-cis-retinal for continued formation of visual pigments and sustained vision. Proteins in the retinal pigment epithelium (RPE), a cell layer adjacent to the photoreceptor outer segments, form the well- established “dark” regeneration pathway known as the classical visual cycle. This pathway is sufficient to maintain continuous rod function and support cone photoreceptors as well although its throughput has to be augmented by additional mechanism(s) to maintain pigment levels in the face of high rates of photon capture. Recent studies indicate that the classical visual cycle works together with light-dependent pro- cesses in both the RPE and neural retina to ensure adequate 11-cis-retinal production under natural illu- minances that can span ten orders of magnitude. Further elucidation of the interplay between these complementary systems is fundamental to understanding how cone-mediated vision is sustained in vivo. Here, we describe recent -
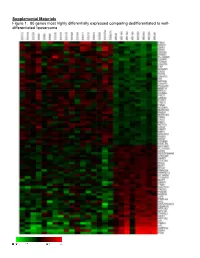
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

Identification of Residues of the H-Ras Protein Critical for Functional Interaction with Guanine Nucleotide Exchange Factors RAYMOND D
MOLECULAR AND CELLULAR BIOLOGY, Feb. 1994, p. 1104-1112 Vol. 14, No. 2 0270-7306/94/$04.00+0 Copyright © 1994, American Society for Microbiology Identification of Residues of the H-Ras Protein Critical for Functional Interaction with Guanine Nucleotide Exchange Factors RAYMOND D. MOSTELLER, JAEWON HAN, AND DANIEL BROEK* Department ofBiochemistry and Molecular Biology, Kenneth Norris Jr. Cancer Hospital and Research Center, University of Southern California School ofMedicine, Los Angeles, California 90033 Received 10 September 1993/Returned for modification 21 October 1993/Accepted 29 October 1993 Ras proteins are activated in vivo by guanine nucleotide exchange factors encoded by genes homologous to the CDC25 gene ofSaccharomyces cerevisiae. We have taken a combined genetic and biochemical approach to probe the sites on Ras proteins important for interaction with such exchange factors and to further probe the mechanism ofCDC25-catalyzed GDP-GTP exchange. Random mutagenesis coupled with genetic selection in S. cerevisiae was used to generate second-site mutations within human H-ras-alalS which could suppress the ability of the Ala-15 substitution to block CDC25 function. We transferred these second-site suppressor mutations to normal H-ras and oncogenic H-rasva1l2 to test whether they induced a general loss of function or whether they selectively affected CDC25 interaction. Four highly selective mutations were discovered, and they affected the surface-located amino acid residues 62, 63, 67, and 69. Two lines of evidence suggested that these residues may be involved in binding to CDC25: (i) using the yeast two-hybrid system, we demonstrated that these mutants cannot bind CDC25 under conditions where the wild-type H-Ras protein can; (ii) we demonstrated that the binding to H-Ras of monoclonal antibody Y13-259, whose epitope has been mapped to residues 63, 65, 66, 67, 70, and 73, is blocked by the mouse sosl and yeast CDC25 gene products. -

Investigating Cone Photoreceptor Development Using Patient-Derived NRL Null Retinal Organoids
ARTICLE https://doi.org/10.1038/s42003-020-0808-5 OPEN Investigating cone photoreceptor development using patient-derived NRL null retinal organoids Alyssa Kallman1,11, Elizabeth E. Capowski 2,11, Jie Wang 3, Aniruddha M. Kaushik4, Alex D. Jansen2, Kimberly L. Edwards2, Liben Chen4, Cynthia A. Berlinicke3, M. Joseph Phillips2,5, Eric A. Pierce6, Jiang Qian3, ✉ ✉ Tza-Huei Wang4,7, David M. Gamm2,5,8 & Donald J. Zack 1,3,9,10 1234567890():,; Photoreceptor loss is a leading cause of blindness, but mechanisms underlying photoreceptor degeneration are not well understood. Treatment strategies would benefit from improved understanding of gene-expression patterns directing photoreceptor development, as many genes are implicated in both development and degeneration. Neural retina leucine zipper (NRL) is critical for rod photoreceptor genesis and degeneration, with NRL mutations known to cause enhanced S-cone syndrome and retinitis pigmentosa. While murine Nrl loss has been characterized, studies of human NRL can identify important insights for human retinal development and disease. We utilized iPSC organoid models of retinal development to molecularly define developmental alterations in a human model of NRL loss. Consistent with the function of NRL in rod fate specification, human retinal organoids lacking NRL develop S- opsin dominant photoreceptor populations. We report generation of two distinct S-opsin expressing populations in NRL null retinal organoids and identify MEF2C as a candidate regulator of cone development. 1 Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, USA. 2 Waisman Center, University of Wisconsin-Madison, Madison, USA. 3 Department of Ophthalmology, Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, USA. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Activation and Molecular Recognition of the GPCR Rhodopsin –
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Institutional Repository of the Freie Universität Berlin Activation and molecular recognition of the GPCR rhodopsin – Insights from time-resolved fluorescence depolarization and single molecule experiments Tai-Yang Kim, Thomas Schlieter, Sebastian Haase, and Ulrike Alexiev Physics Department, Freie Universität Berlin, Arnimallee 14, D-14195 Berlin, Germany Correspondence: Dr. Ulrike Alexiev, Physics Department, Freie Universität Berlin, Arnimallee 14, D-14195 Berlin, Germany, Phone: +49-30-838-55157, Fax: +49-30-838-56150, email: [email protected] Keywords: GPCR, visual phototransduction, rhodopsin, GPCR helix 8, protein dynamics, time-resolved fluorescence depolarization, fluorescence anisotropy, surface potential, single particle tracking 1 Abstract The cytoplasmic surface of the G-protein coupled receptor (GPCR) rhodopsin is a key element in membrane receptor activation, molecular recognition by signaling molecules, and receptor deactivation. Understanding of the coupling between conformational changes in the intramembrane domain and the membrane-exposed surface of the photoreceptor rhodopsin is crucial for the elucidation of molecular mechanism in GPCR activation. As little is known about protein dynamics, particularly the conformational dynamics of the cytoplasmic surface elements on the nanoseconds timescale, we utilized time-resolved fluorescence anisotropy experiments and site-directed fluorescence labeling to provide information on both, conformational space and motion. We summarize our recent advances in understanding rhodopsin dynamics and function using time-resolved fluorescence depolarization and single molecule fluorescence experiments, with particular focus on the amphipathic helix 8, lying parallel to the cytoplasmic membrane surface and connecting transmembrane helix 7 with the long C-terminal tail. -

G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision
The Journal of Neuroscience, June 1, 2011 • 31(22):8067–8077 • 8067 Cellular/Molecular G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision Alexander V. Kolesnikov,1 Loryn Rikimaru,2 Anne K. Hennig,1 Peter D. Lukasiewicz,1 Steven J. Fliesler,4,5,6,7 Victor I. Govardovskii,8 Vladimir J. Kefalov,1 and Oleg G. Kisselev2,3 1Department of Ophthalmology and Visual Sciences, Washington University School of Medicine, St. Louis, Missouri 63110, Departments of 2Ophthalmology and 3Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, Missouri 63104, 4Research Service, Veterans Administration Western New York Healthcare System, and Departments of 5Ophthalmology (Ross Eye Institute) and 6Biochemistry, University at Buffalo/The State University of New York (SUNY), and 7SUNY Eye Institute, Buffalo, New York 14215, and 8Sechenov Institute for Evolutionary Physiology and Biochemistry, Russian Academy of Sciences, Saint Petersburg 194223, Russia A fundamental question of cell signaling biology is how faint external signals produce robust physiological responses. One universal mechanism relies on signal amplification via intracellular cascades mediated by heterotrimeric G-proteins. This high amplification system allows retinal rod photoreceptors to detect single photons of light. Although much is now known about the role of the ␣-subunit of the rod-specific G-protein transducin in phototransduction, the physiological function of the auxiliary ␥-complex in this process remains a mystery. Here, we show that elimination of the transducin ␥-subunit drastically reduces signal amplification in intact mouse rods. The consequence is a striking decline in rod visual sensitivity and severe impairment of nocturnal vision. Our findings demonstrate that transducin ␥-complex controls signal amplification of the rod phototransduction cascade and is critical for the ability of rod photoreceptors to function in low light conditions. -

G Protein-Coupled Receptors
www.aladdin-e.com Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: [email protected] Email EU: [email protected] Email Asia Pacific: [email protected] G PROTEIN-COUPLED RECEPTORS Overview: The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20–24 hydrophobic amino acids arranged as a-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Fredriksson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor (see Page S195) has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. -

Comparative Analysis of Human Chromosome 7Q21 and Mouse
Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing David Monk,1,6 Alexandre Wagschal,2 Philippe Arnaud,2 Pari-Sima Mu¨ller,3 Layla Parker-Katiraee,4 Déborah Bourc’his,5 Stephen W. Scherer,4 Robert Feil,2 Philip Stanier,1 and Gudrun E. Moore1 1Institute of Child Health, London WC1N 1EH, United Kingdom; 2Institute of Molecular Genetics, CNRS UMR-5535 and University of Montpellier-II, 34293 Montpellier, France; 3Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, United Kingdom; 4Center for Applied Genomics, The Hospital for Sick Children, Toronto M5G 1L7, Canada; 5Inserm U741, F-75251 Paris Cedex 05, France Genomic imprinting is a developmentally important mechanism that involves both differential DNA methylation and allelic histone modifications. Through detailed comparative characterization, a large imprinted domain mapping to chromosome 7q21 in humans and proximal chromosome 6 in mice was redefined. This domain is organized around a maternally methylated CpG island comprising the promoters of the adjacent PEG10 and SGCE imprinted genes. Examination of Dnmt3l−/+ conceptuses shows that imprinted expression for all genes of the cluster depends upon the germline methylation at this putative “imprinting control region” (ICR). Similarly as for other ICRs, we find its DNA-methylated allele to be associated with trimethylation of lysine 9 on histone H3 (H3K9me3) and trimethylation of lysine 20 on histone H4 (H4K20me3), whereas the transcriptionally active paternal allele is enriched in H3K4me2 and H3K9 acetylation. -

WO 2011/043591 A2 14 April 2011 (14.04.2011) PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau I (10) International Publication Number (43) International Publication Date WO 2011/043591 A2 14 April 2011 (14.04.2011) PCT (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12N 5/071 (2010.01) C12N 5/02 (2006.01) kind of national protection available): AE, AG, AL, AM, C12N 5/07 (2010.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) Number: International Application DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/KR20 10/006832 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 6 October 2010 (06.10.2010) MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, SE, (25) Filing Language: English SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, (26) Publication Language: English TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 10-2009-0094854 6 October 2009 (06. 10.2009) KR kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, (71) Applicant (for all designated States except US): SNU ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, R&DB FOUNDATION [KR/KR]; San 56-1, Sillim- TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, dong, Gwanak-gu, Seoul 15 1-742 (KR).