Personalized Diagnostics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Deubiquitinases in Cancer: New Functions and Therapeutic Options
Oncogene (2012) 31, 2373–2388 & 2012 Macmillan Publishers Limited All rights reserved 0950-9232/12 www.nature.com/onc REVIEW Deubiquitinases in cancer: new functions and therapeutic options JM Fraile1, V Quesada1, D Rodrı´guez, JMP Freije and C Lo´pez-Otı´n Departamento de Bioquı´mica y Biologı´a Molecular, Facultad de Medicina, Instituto Universitario de Oncologı´a, Universidad de Oviedo, Oviedo, Spain Deubiquitinases (DUBs) have fundamental roles in the Hunter, 2010). Consistent with the functional relevance ubiquitin system through their ability to specifically of proteases in these processes, alterations in their deconjugate ubiquitin from targeted proteins. The human structure or in the mechanisms controlling their genome encodes at least 98 DUBs, which can be grouped spatiotemporal expression patterns and activities cause into 6 families, reflecting the need for specificity in diverse pathologies such as arthritis, neurodegenerative their function. The activity of these enzymes affects the alterations, cardiovascular diseases and cancer. Accord- turnover rate, activation, recycling and localization ingly, many proteases are an important focus of of multiple proteins, which in turn is essential for attention for the pharmaceutical industry either as drug cell homeostasis, protein stability and a wide range of targets or as diagnostic and prognostic biomarkers signaling pathways. Consistent with this, altered DUB (Turk, 2006; Drag and Salvesen, 2010). function has been related to several diseases, including The recent availability of the genome sequence cancer. Thus, multiple DUBs have been classified as of different organisms has facilitated the identification oncogenes or tumor suppressors because of their regula- of their entire protease repertoire, which has been tory functions on the activity of other proteins involved in defined as degradome (Lopez-Otin and Overall, 2002). -

Supplementary Material Contents
Supplementary Material Contents Immune modulating proteins identified from exosomal samples.....................................................................2 Figure S1: Overlap between exosomal and soluble proteomes.................................................................................... 4 Bacterial strains:..............................................................................................................................................4 Figure S2: Variability between subjects of effects of exosomes on BL21-lux growth.................................................... 5 Figure S3: Early effects of exosomes on growth of BL21 E. coli .................................................................................... 5 Figure S4: Exosomal Lysis............................................................................................................................................ 6 Figure S5: Effect of pH on exosomal action.................................................................................................................. 7 Figure S6: Effect of exosomes on growth of UPEC (pH = 6.5) suspended in exosome-depleted urine supernatant ....... 8 Effective exosomal concentration....................................................................................................................8 Figure S7: Sample constitution for luminometry experiments..................................................................................... 8 Figure S8: Determining effective concentration ......................................................................................................... -

The Proteasomal Deubiquitinating Enzyme PSMD14 Regulates Macroautophagy by Controlling Golgi-To-ER Retrograde Transport
Supplementary Materials The proteasomal deubiquitinating enzyme PSMD14 regulates macroautophagy by controlling Golgi-to-ER retrograde transport Bustamante HA., et al. Figure S1. siRNA sequences directed against human PSMD14 used for Validation Stage. Figure S2. Primer pairs sequences used for RT-qPCR. Figure S3. The PSMD14 DUB inhibitor CZM increases the Golgi apparatus area. Immunofluorescence microscopy analysis of the Golgi area in parental H4 cells treated for 4 h either with the vehicle (DMSO; Control) or CZM. The Golgi marker GM130 was used to determine the region of interest in each condition. Statistical significance was determined by Student's t-test. Bars represent the mean ± SEM (n =43 cells). ***P <0.001. Figure S4. CZM causes the accumulation of KDELR1-GFP at the Golgi apparatus. HeLa cells expressing KDELR1-GFP were either left untreated or treated with CZM for 30, 60 or 90 min. Cells were fixed and representative confocal images were acquired. Figure S5. Effect of CZM on proteasome activity. Parental H4 cells were treated either with the vehicle (DMSO; Control), CZM or MG132, for 90 min. Protein extracts were used to measure in vitro the Chymotrypsin-like peptidase activity of the proteasome. The enzymatic activity was quantified according to the cleavage of the fluorogenic substrate Suc-LLVY-AMC to AMC, and normalized to that of control cells. The statistical significance was determined by One-Way ANOVA, followed by Tukey’s test. Bars represent the mean ± SD of biological replicates (n=3). **P <0.01; n.s., not significant. Figure S6. Effect of CZM and MG132 on basal macroautophagy. (A) Immunofluorescence microscopy analysis of the subcellular localization of LC3 in parental H4 cells treated with either with the vehicle (DMSO; Control), CZM for 4 h or MG132 for 6 h. -

Supplementary Data ASXL2 Regulates Hematopoiesis in Mice and Its
Supplementary data ASXL2 regulates hematopoiesis in mice and its deficiency promotes myeloid expansion Supplementary Methods Genomic DNA extraction Genomic DNA was extracted from BM mononuclear cells using a DNA extraction kit (Puregene Gentra System, Minneapolis, MN, USA) according to the manufacturer’s instructions. Genomic DNA was quantified using Qubit Fluorometer (Life Technologies) and DNA integrity was assessed by agarose gel electrophoresis. For samples with low quantity, DNA was amplified using REPLI-g Ultrafast mini kit (Qiagen). Peripheral blood analysis Complete peripheral blood counts were analysed using Abbott Cell-Dyn 3700 hematology analyzer (Abbott Laboratories). Expression analysis of Asxl2 and Asxl1 Transcript levels of Asxl2 and Asxl1 were estimated using quantitative RT-PCR with following primers: Asxl2 primer set 1, ATTCGACAAGAGATTGAGAAGGAG (forward) and TTTCTGTGAATCTTCAAGGCTTAG (reverse); Asxl2 primer set 2, GCCCTTAACAATGAGTTCTTCACT (forward) and TCCACAGCTCTACTTTCTTCTCCT (reverse); Asxl1 primers, GGTGGAACAATGGAAGGAAA (forward) and CTGGCCGAGAACGTTTCTTA (reverse). Asxl2 protein was detected in immunoblot using anti-ASXL2 antibody (Bethyl). In vitro differentiation of bone marrow cells Bone marrow (BM) cells were cultured in IMDM containing 20% FBS and 10 ng/ml IL3, 10 ng/ml IL6, 20 ng/ml SCF and 20 ng/ml GMCSF for two weeks. For FACS analysis, cells were washed, stained with fluorochrome-conjugated antibodies and analysed on FACS LSR II flow cytometer (BD Biosciences) using FACSDIVA software (BD Biosciences). Colony re-plating assay Bone marrow cells were plated in methylcellulose medium containing mouse stem cell factor (SCF), mouse interleukin 3 (IL-3), human interleukin 6 (IL-6) and human erythropoietin (MethoCult GF M3434; StemCell Technologies). Colonies were enumerated after 9-12 days and cells were harvested for re-plating. -

For Personal Use. Only Reproduce with Permission from the Lancet
Correlation to non-HGNT Accesion group 2 Description AA897204 1.479917 ESTs T85111 1.286576 null T60168 | AI821353 1.274487 thyroid transcription factor 1 AA600173 1.183065 ubiquitin-conjugating enzyme E2A (RAD6 homolog) R55745 1.169339 ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) AA400492 1.114935 ESTs AA864791 1.088826 hypothetical protein FLJ21313 N53758 1.070402 EST AI216628 1.067763 ESTs AI167637 1.058561 ESTs AA478265 1.056331 similar to transmembrane receptor Unc5H1 AA969924 1.039315 EST AI074650 1.039043 hypothetical protein FLJ13842 R20763 1.035807 ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) AI347081 1.034518 Ca2+-dependent activator protein for secretion R44386 1.028005 ESTs AA976699 1.027227 chromogranin A (parathyroid secretory protein 1) AA634475 1.026766 KIAA1796 protein AA496809 1.02432 SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 H16572 1.013059 null H29013 1.002117 seizure related 6 homolog (mouse)-like AI299731 1.001053 Homo sapiens nanos mRNA, partial cds AA400194 0.9950039 EST AI216537 | AI820870 0.9737153 Homo sapiens cDNA FLJ39674 fis, clone SMINT2009505 AA426408 0.9728649 type I transmembrane receptor (seizure-related protein) AA971742 | AI733380 0.9707561 achaete-scute complex-like 1 (Drosophila) R41450 0.9655133 ESTs AA487505 0.9636143 immunoglobulin superfamily, member 4 AA404337 0.957686 thymus high mobility group box protein TOX N68578 0.9552571 ESTs R45008 0.9422938 ELAV (embryonic lethal, abnormal -

Deciphering Mechanisms of Diapause Entry and Exit in the Annual Killifish Austrofundulus Limnaeus
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses 4-15-2021 Connecting Environment and Phenotype: Deciphering Mechanisms of Diapause Entry and Exit in the Annual Killifish Austrofundulus limnaeus Erin Monica Davis Portland State University Follow this and additional works at: https://pdxscholar.library.pdx.edu/open_access_etds Part of the Animal Sciences Commons, and the Biology Commons Let us know how access to this document benefits ou.y Recommended Citation Davis, Erin Monica, "Connecting Environment and Phenotype: Deciphering Mechanisms of Diapause Entry and Exit in the Annual Killifish Austrofundulus limnaeus" (2021). Dissertations and Theses. Paper 5680. https://doi.org/10.15760/etd.7552 This Thesis is brought to you for free and open access. It has been accepted for inclusion in Dissertations and Theses by an authorized administrator of PDXScholar. Please contact us if we can make this document more accessible: [email protected]. Connecting Environment and Phenotype: Deciphering Mechanisms of Diapause Entry and Exit in the Annual Killifish Austrofundulus limnaeus by Erin Monica Davis A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Biology Thesis Committee: Jason Podrabsky, Chair Deborah Duffield Annie Lindgren Portland State University 2021 ©2021 Erin Monica Davis ABSTRACT Embryonic development is complex, dynamic, and dependent on environmental factors. Mechanisms of sensing and integrating environmental stimuli are diverse, and understanding these mechanisms in extant species can elucidate how complex phenotypes emerge from genomic information expressed in an environmental context. In Austrofundulus limnaeus, an annual killifish with alternative developmental trajectories, light and temperature are vital factors that determine if an embryo will enter a state of diapause. -

Comparative Analysis of the Ubiquitin-Proteasome System in Homo Sapiens and Saccharomyces Cerevisiae
Comparative Analysis of the Ubiquitin-proteasome system in Homo sapiens and Saccharomyces cerevisiae Inaugural-Dissertation zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Universität zu Köln vorgelegt von Hartmut Scheel aus Rheinbach Köln, 2005 Berichterstatter: Prof. Dr. R. Jürgen Dohmen Prof. Dr. Thomas Langer Dr. Kay Hofmann Tag der mündlichen Prüfung: 18.07.2005 Zusammenfassung I Zusammenfassung Das Ubiquitin-Proteasom System (UPS) stellt den wichtigsten Abbauweg für intrazelluläre Proteine in eukaryotischen Zellen dar. Das abzubauende Protein wird zunächst über eine Enzym-Kaskade mit einer kovalent gebundenen Ubiquitinkette markiert. Anschließend wird das konjugierte Substrat vom Proteasom erkannt und proteolytisch gespalten. Ubiquitin besitzt eine Reihe von Homologen, die ebenfalls posttranslational an Proteine gekoppelt werden können, wie z.B. SUMO und NEDD8. Die hierbei verwendeten Aktivierungs- und Konjugations-Kaskaden sind vollständig analog zu der des Ubiquitin- Systems. Es ist charakteristisch für das UPS, daß sich die Vielzahl der daran beteiligten Proteine aus nur wenigen Proteinfamilien rekrutiert, die durch gemeinsame, funktionale Homologiedomänen gekennzeichnet sind. Einige dieser funktionalen Domänen sind auch in den Modifikations-Systemen der Ubiquitin-Homologen zu finden, jedoch verfügen diese Systeme zusätzlich über spezifische Domänentypen. Homologiedomänen lassen sich als mathematische Modelle in Form von Domänen- deskriptoren (Profile) beschreiben. Diese Deskriptoren können wiederum dazu verwendet werden, mit Hilfe geeigneter Verfahren eine gegebene Proteinsequenz auf das Vorliegen von entsprechenden Homologiedomänen zu untersuchen. Da die im UPS involvierten Homologie- domänen fast ausschließlich auf dieses System und seine Analoga beschränkt sind, können domänen-spezifische Profile zur Katalogisierung der UPS-relevanten Proteine einer Spezies verwendet werden. Auf dieser Basis können dann die entsprechenden UPS-Repertoires verschiedener Spezies miteinander verglichen werden. -

BMC Biochemistry Biomed Central
BMC Biochemistry BioMed Central Review Open Access Deubiquitylating enzymes and disease Shweta Singhal1, Matthew C Taylor2 and Rohan T Baker*1 Address: 1Ubiquitin Laboratory, Division of Molecular Bioscience, The John Curtin School of Medical Research, The Australian National University, Canberra, ACT 0200, Australia and 2CSIRO Entomology, GPO Box 1700, Canberra, ACT 2601, Australia Email: Shweta Singhal - [email protected]; Matthew C Taylor - [email protected]; Rohan T Baker* - [email protected] * Corresponding author Published: 21 October 2008 <supplement> <title> <p>Ubiquitin-Proteasome System in Disease Part 2</p> </title> <editor>John Mayer and Rob Layfield</editor> <note>Reviews</note> </supplement> BMC Biochemistry 2008, 9(Suppl 1):S3 doi:10.1186/1471-2091-9-S1-S3 This article is available from: http://www.biomedcentral.com/1471-2091/9/S1/S3 © 2008 Singhal et al; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Deubiquitylating enzymes (DUBs) can hydrolyze a peptide, amide, ester or thiolester bond at the C-terminus of UBIQ (ubiquitin), including the post-translationally formed branched peptide bonds in mono- or multi-ubiquitylated conjugates. DUBs thus have the potential to regulate any UBIQ- mediated cellular process, the two best characterized being proteolysis and protein trafficking. Mammals contain some 80–90 DUBs in five different subfamilies, only a handful of which have been characterized with respect to the proteins that they interact with and deubiquitylate. -

Bone and Soft Tissue Pathology Specimens and 127 Surgicals (Biopsies and Resections)
ANNUAL MEETING ABSTRACTS 11A the morphologic spectrum and etiology of DAD encountered during adult autopsy in (non-EWSR1) NR4A3 gene fusions (TAF15, TCF12) showed distinctive plasmacytoid an inner city teaching hospital. The diagnostic utility of post mortem lung culture was / rhabdoid morphology, with increased cellularity, cytologic atypia and high mitotic also evaluated. counts. Follow-up showed that only 1 of 16 patients with EWSR1-rearranged tumors Design: A retrospective study was performed on all adult autopsies from July 2010 to died of disease, in contrast to 3 of 7 (43%) patients with TAF15–rearranged tumors. July 2013 with fi nal histopathologic diagnosis of DAD. The histopathological features Conclusions: In conclusion, EMCs with variant NR4A3 gene fusions show a higher of DAD were re-evaluated by one autopsy pathologist and one pathology resident, incidence of rhabdoid phenotype, high grade morphology and a more aggressive based on the duration (exudative or proliferative phase), severity (bilateral/unilateral; outcome compared to the more common EWSR1-NR4A3 positive tumors. Furthermore, focal/extensive) and pattern (classical vs. acute fi brinous and organizing pneumonia as EWSR1 FISH break-apart assay is the preferred ancillary test to confi rm diagnosis aka AFOP). Clinical history, pre and post mortem laboratory investigations, including of EMC, tumors with variant NR4A3 gene fusions remain under-recognized and often postmortem lung culture (for bacteria, mycobacteria, fungi and virus) were reviewed misdiagnosed. FISH assay for NR4A3 rearrangements recognizes >95% of EMCs and to elucidate etiology. should be an additional tool in EWSR1-negative tumors. Results: 36 (16.2 %) cases showed histopathologic features of DAD out of 222 adult autopsies in the three year study period. -
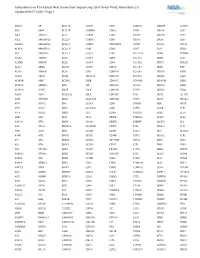
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1 ABCC3 AR BCL11A CANT1 CDK1 CMKLR1 DAB2IP DUSP9 ABI1 ARAF BCL11B CAPRIN1 CDK12 CNBP DACH1 E2F1 ABL1 ARFRP1 BCL2 CAPZB CDK2 CNOT2 DACH2 E2F3 ABL2 ARHGAP20 BCL2A1 CARD11 CDK4 CNTN1 DAXX EBF1 ABLIM1 ARHGAP26 BCL2L1 CARM1 CDK5RAP2 CNTRL DCLK2 ECT2L ACACA ARHGEF12 BCL2L11 CARS CDK6 COG5 DCN EDIL3 ACE ARHGEF7 BCL2L2 CASC5 CDK7 COL11A1 DDB1 EDNRB ACER1 ARID1A BCL3 CASP3 CDK8 COL1A1 DDB2 EED ACSBG1 ARID1B BCL6 CASP7 CDK9 COL1A2 DDIT3 EEFSEC ACSL3 ARID2 BCL7A CASP8 CDKL5 COL3A1 DDR2 EGF ACSL6 ARID5B BCL9 CAV1 CDKN1A COL6A3 DDX10 EGFR ACVR1 ARIH2 BCOR CBFA2T3 CDKN1B COL9A3 DDX20 EGR1 ACVR1B ARNT BCORL1 CBFB CDKN1C COMMD1 DDX39B EGR2 ACVR1C ARRDC4 BCR CBL CDKN2A COX6C DDX3X EGR3 ACVR2A ASMTL BDNF CBLB CDKN2B CPNE1 DDX41 EGR4 ADD3 ASPH BHLHE22 CBLC CDKN2C CPS1 DDX5 EIF1AX ADM ASPSCR1 BICC1 CCDC28A CDKN2D CPSF6 DDX6 EIF4A2 AFF1 ASTN2 BIN1 CCDC6 CDX1 CRADD DEK EIF4E AFF3 ASXL1 BIRC3 CCDC88C CDX2 CREB1 DGKB ELF3 AFF4 ASXL2 BIRC6 CCK CEBPA CREB3L1 DGKI ELF4 AGR3 ATF1 BLM CCL2 CEBPB CREB3L2 DGKZ ELK4 AHCYL1 ATF3 BMP4 CCNA2 CEBPD CREBBP DICER1 ELL AHI1 ATG13 BMPR1A CCNB1IP1 CEBPE CRKL DIRAS3 ELN AHR ATG5 BRAF CCNB3 CENPF CRLF2 DIS3 ELOVL2 AHRR ATIC BRCA1 CCND1 CENPU CRTC1 DIS3L2 ELP2 AIP ATL1 BRCA2 CCND2 CEP170B CRTC3 DKK1 EML1 AK2 ATM BRCC3 CCND3 CEP57 CSF1 DKK2 EML4 AK5 ATP1B4 BRD1 CCNE1 CEP85L CSF1R DKK4 ENPP2 AKAP12 ATP8A2 BRD3 CCNG1 CHCHD7 CSF3 DLEC1 EP300 AKAP6 ATR BRD4 CCT6B CHD2 CSF3R DLL1 EP400 AKAP9 ATRNL1 BRIP1 CD19 CHD4 CSNK1A1 DLL3 -

Deubiquitylation of Deubiquitylases.Pdf
Deubiquitylation of deubiquitylases Saba Haq1 and Suresh Ramakrishna2,3 rsob.royalsocietypublishing.org 1Department of Life Science, College of Natural Sciences, 2Graduate School of Biomedical Science and Engineering, and 3College of Medicine, Hanyang University, Seoul, South Korea SR, 0000-0002-4038-1085 Deubiquitylating enzymes (DUBs) reverse the ubiquitylation of target pro- teins, thereby regulating diverse cellular functions. In contrast to the Review plethora of research being conducted on the ability of DUBs to counter the degradation of cellular proteins or auto-ubiquitylated E3 ligases, very little Cite this article: Haq S, Ramakrishna S. 2017 is known about the mechanisms of DUB regulation. In this review paper, Deubiquitylation of deubiquitylases. Open Biol. we summarize a novel possible mechanism of DUB deubiquitylation by 7: 170016. other DUBs. The available data suggest the need for further experiments http://dx.doi.org/10.1098/rsob.170016 to validate and characterize this notion of ‘Dubbing DUBs’. The current studies indicate that the idea of deubiquitylation of DUBs by other DUBs is still in its infancy. Nevertheless, future research holds the promise of validation of this concept. Received: 17 January 2017 Accepted: 30 April 2017 1. Introduction The ubiquitin proteasome pathway, prominently responsible for the targeted degradation of usually short-lived proteins (e.g. cell-cycle regulatory proteins), Subject Area: is a major part of various cellular regulation events. Two decades ago, Ye biotechnology/molecular biology/cellular et al. uncovered three enzymes required for this process and termed them biology ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin ligase (E3) [1]. E1 activates ubiquitin by producing a ubiquitin- Keywords: adenylate intermediate, followed by subsequent transfer of ubiquitin to an auto-regulation, deubiquitylating DUBs, active site cysteine residue.