Deciphering Mechanisms of Diapause Entry and Exit in the Annual Killifish Austrofundulus Limnaeus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Roles of Id3 and IL-13 in a Mouse Model of Autoimmune Exocrinopathy
Roles of Id3 and IL-13 in a Mouse Model of Autoimmune Exocrinopathy by Ian Lawrence Belle Department of Immunology Duke University Date:_______________________ Approved: ___________________________ Yuan Zhuang, Supervisor ___________________________ Michael Krangel, Chair ___________________________ Qi-jing Li ___________________________ Richard Lee Reinhardt ___________________________ Arno Greenleaf Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Immunology in the Graduate School of Duke University 2015 ABSTRACT Roles of Id3 and IL-13 in a Mouse Model of Autoimmune Exocrinopathy by Ian Lawrence Belle Department of Immunology Duke University Date:_______________________ Approved: ___________________________ Yuan Zhuang, Supervisor ___________________________ Michael Krangel, Chair ___________________________ Qi-jing Li ___________________________ Richard Lee Reinhardt ___________________________ Arno Greenleaf An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Immunology in the Graduate School of Duke University 2015 Copyright by Ian Lawrence Belle 2015 Abstract Within the field of immunology, the existence of autoimmune diseases presents a unique set of challenges. The immune system typically protects the host by identifying foreign pathogens and mounting an appropriate response to eliminate them. Great strides have been made in understanding how foreign pathogens are identified and responded to, leading to the development of powerful immunological tools, such as vaccines and a myriad of models used to study infectious diseases and processes. However, it is occasionally possible for host tissues themselves to be inappropriately identified as foreign, prompting an immune response that attempts to eliminate the host tissue. The immune system has processes in place, referred to as selection, designed to prevent the development of cells capable of recognizing the self as foreign. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Deubiquitinases in Cancer: New Functions and Therapeutic Options
Oncogene (2012) 31, 2373–2388 & 2012 Macmillan Publishers Limited All rights reserved 0950-9232/12 www.nature.com/onc REVIEW Deubiquitinases in cancer: new functions and therapeutic options JM Fraile1, V Quesada1, D Rodrı´guez, JMP Freije and C Lo´pez-Otı´n Departamento de Bioquı´mica y Biologı´a Molecular, Facultad de Medicina, Instituto Universitario de Oncologı´a, Universidad de Oviedo, Oviedo, Spain Deubiquitinases (DUBs) have fundamental roles in the Hunter, 2010). Consistent with the functional relevance ubiquitin system through their ability to specifically of proteases in these processes, alterations in their deconjugate ubiquitin from targeted proteins. The human structure or in the mechanisms controlling their genome encodes at least 98 DUBs, which can be grouped spatiotemporal expression patterns and activities cause into 6 families, reflecting the need for specificity in diverse pathologies such as arthritis, neurodegenerative their function. The activity of these enzymes affects the alterations, cardiovascular diseases and cancer. Accord- turnover rate, activation, recycling and localization ingly, many proteases are an important focus of of multiple proteins, which in turn is essential for attention for the pharmaceutical industry either as drug cell homeostasis, protein stability and a wide range of targets or as diagnostic and prognostic biomarkers signaling pathways. Consistent with this, altered DUB (Turk, 2006; Drag and Salvesen, 2010). function has been related to several diseases, including The recent availability of the genome sequence cancer. Thus, multiple DUBs have been classified as of different organisms has facilitated the identification oncogenes or tumor suppressors because of their regula- of their entire protease repertoire, which has been tory functions on the activity of other proteins involved in defined as degradome (Lopez-Otin and Overall, 2002). -

Based Mechanism Study on the Beneficial Effects of Vitamin D
www.nature.com/scientificreports OPEN Network Systems Pharmacology- Based Mechanism Study on the Benefcial Efects of Vitamin D against Psychosis in Alzheimer’s Disease Peihao Fan1, Xiguang Qi1, Robert A. Sweet2,3* & Lirong Wang1* Alzheimer’s disease (AD) is a chronic neurodegenerative disease with signifcant fnancial costs and negative impacts on quality of life. Psychotic symptoms, i.e., the presence of delusions and/ or hallucinations, is a frequent complication of AD. About 50% of AD patients will develop psychotic symptoms (AD with Psychosis, or AD + P) and these patients will experience an even more rapid cognitive decline than AD patients without psychosis (AD-P). In a previous analysis on medication records of 776 AD patients, we had shown that use of Vitamin D was associated with delayed time to psychosis in AD patients and Vitamin D was used more by AD-P than AD + P patients. To explore the potential molecular mechanism behind our fndings, we applied systems pharmacology approaches to investigate the crosstalk between AD and psychosis. Specifcally, we built protein-protein interaction (PPI) networks with proteins encoded by AD- and psychosis-related genes and Vitamin D-perturbed genes. Using network analysis we identifed several high-impact genes, including NOTCH4, COMT, CACNA1C and DRD3 which are related to calcium homeostasis. The new fndings highlight the key role of calcium-related signaling pathways in AD + P development and may provide a new direction and facilitate hypothesis generation for future drug development. Alzheimer’s disease (AD) is a chronic neurodegenerative disease commonly seen in the aging population, and the presence of AD is responsible for a signifcant decrease in the quality of life1. -

SUPPLEMENTARY MATERIAL Bone Morphogenetic Protein 4 Promotes
www.intjdevbiol.com doi: 10.1387/ijdb.160040mk SUPPLEMENTARY MATERIAL corresponding to: Bone morphogenetic protein 4 promotes craniofacial neural crest induction from human pluripotent stem cells SUMIYO MIMURA, MIKA SUGA, KAORI OKADA, MASAKI KINEHARA, HIROKI NIKAWA and MIHO K. FURUE* *Address correspondence to: Miho Kusuda Furue. Laboratory of Stem Cell Cultures, National Institutes of Biomedical Innovation, Health and Nutrition, 7-6-8, Saito-Asagi, Ibaraki, Osaka 567-0085, Japan. Tel: 81-72-641-9819. Fax: 81-72-641-9812. E-mail: [email protected] Full text for this paper is available at: http://dx.doi.org/10.1387/ijdb.160040mk TABLE S1 PRIMER LIST FOR QRT-PCR Gene forward reverse AP2α AATTTCTCAACCGACAACATT ATCTGTTTTGTAGCCAGGAGC CDX2 CTGGAGCTGGAGAAGGAGTTTC ATTTTAACCTGCCTCTCAGAGAGC DLX1 AGTTTGCAGTTGCAGGCTTT CCCTGCTTCATCAGCTTCTT FOXD3 CAGCGGTTCGGCGGGAGG TGAGTGAGAGGTTGTGGCGGATG GAPDH CAAAGTTGTCATGGATGACC CCATGGAGAAGGCTGGGG MSX1 GGATCAGACTTCGGAGAGTGAACT GCCTTCCCTTTAACCCTCACA NANOG TGAACCTCAGCTACAAACAG TGGTGGTAGGAAGAGTAAAG OCT4 GACAGGGGGAGGGGAGGAGCTAGG CTTCCCTCCAACCAGTTGCCCCAAA PAX3 TTGCAATGGCCTCTCAC AGGGGAGAGCGCGTAATC PAX6 GTCCATCTTTGCTTGGGAAA TAGCCAGGTTGCGAAGAACT p75 TCATCCCTGTCTATTGCTCCA TGTTCTGCTTGCAGCTGTTC SOX9 AATGGAGCAGCGAAATCAAC CAGAGAGATTTAGCACACTGATC SOX10 GACCAGTACCCGCACCTG CGCTTGTCACTTTCGTTCAG Suppl. Fig. S1. Comparison of the gene expression profiles of the ES cells and the cells induced by NC and NC-B condition. Scatter plots compares the normalized expression of every gene on the array (refer to Table S3). The central line -
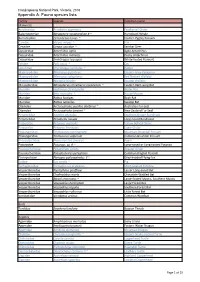
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

A Conserved Tissue-Specific Homeodomain-Less Isoform of MEIS1 Is Downregulated in Colorectal Cancer
Thomas Jefferson University Jefferson Digital Commons Department of Microbiology and Immunology Faculty Papers Department of Microbiology and Immunology 8-17-2011 A conserved tissue-specific homeodomain-less isoform of MEIS1 is downregulated in colorectal cancer. Richard C Crist Department of Microbiology and Immunology, Thomas Jefferson University, Philadelphia Jacquelyn J Roth Department of Microbiology and Immunology, Thomas Jefferson University, Philadelphia Scott A Waldman Department of Pharmacology and Experimental Therapeutics, Thomas Jefferson University, Philadelphia Arthur M Buchberg Department of Microbiology and Immunology, Thomas Jefferson University, Philadelphia Follow this and additional works at: https://jdc.jefferson.edu/mifp Part of the Gastroenterology Commons, Medical Genetics Commons, Medical Microbiology Commons, Oncology Commons, and the Pathology Commons Let us know how access to this document benefits ouy Recommended Citation Crist, Richard C; Roth, Jacquelyn J; Waldman, Scott A; and Buchberg, Arthur M, "A conserved tissue-specific homeodomain-less isoform of MEIS1 is downregulated in colorectal cancer." (2011). Department of Microbiology and Immunology Faculty Papers. Paper 25. https://jdc.jefferson.edu/mifp/25 This Article is brought to you for free and open access by the Jefferson Digital Commons. The Jefferson Digital Commons is a service of Thomas Jefferson University's Center for Teaching and Learning (CTL). The Commons is a showcase for Jefferson books and journals, peer-reviewed scholarly publications, unique historical collections from the University archives, and teaching tools. The Jefferson Digital Commons allows researchers and interested readers anywhere in the world to learn about and keep up to date with Jefferson scholarship. This article has been accepted for inclusion in Department of Microbiology and Immunology Faculty Papers by an authorized administrator of the Jefferson Digital Commons. -

Personalized Diagnostics
The Center for PERSONALIZED DIAGNOSTICS Precision Diagnostics for Personalized Medicine PennMedicine.org/CPD | 215.615.3966 The Center for Personalized Diagnostics (CPD) is a joint initiative between Penn Medicine’s Department of Pathology and Laboratory Medicine and the Abramson Cancer Center. The Center integrates molecular genetics, pathology informatics and genomic pathology to develop personalized diagnostic profiles for individuals with cancer. The CPD offers the highest volume of genome testing in the region. PENNSEQTM HEMATOLOGIC MALIGNANCIES PANEL ABL1 CD79A FANCA IKZF1 NOTCH1 RIT1 TERT ASXL1 CD79B FANCC IL7R NOTCH2 RPS15 TET2 ATM CDKN2A FANCD2 JAK2 NPM1 RRAGC TNFAIP3 B2M CEBPA FANCE JAK3 NRAS RUNX1 TNFRSF14 BCL2 CIITA FANCF KIT PALB2 SETBP1 TP53 BCOR CREBBP FANCG KLF2 PDGFRA SF1 TPMT BCORL1 CSF1R FANCL KLHL6 PHF6 SF3A1 TRAF3 BIRC3 CSF3R FANCM KRAS PLCG1 SF3B1 U2AF1 BRAF CXCR4 FBXW7 MAP2K1 PLCG2 SLX4 U2AF2 BRCA1 DDX3X FLT3 MAPK1 POT1 SMC1A WT1 BRCA2 DDX41 GATA2 MIR142 PRPF40B SOCS1 XPO1 BRINP3 DICER1 GNA13 MPL PTEN SRSF2 XRCC2 BRIP1 DNMT3A GNAS MYC PTPN11 STAG2 ZMYM3 BTK EGR2 HNRNPK MYCN RAD21 STAT3 ZRSR2 CALR ERCC4 ID3 MYD88 RAD51 STAT5B CARD11 ETV6 IDH1 NF1 RAD51C TBL1XR1 CBL EZH2 IDH2 NFKBIE RHOA TCF3 • Accepted Specimens: Blood; bone marrow; formalin-fixed, paraffin-embedded (FFPE) • Detects: Single nucleotide variants (SNVs); small indels; copy number gains in ABL1, tissue; fresh tissue in PreservCyt PDGFRA, and MYC • Minimum Requirements: 10% tumor nuclei for tissue, 100ng of DNA (non-FFPE), 200ng • Limitations: Lower limit -

Association Between Vitamin D Receptor Rs731236 (Taq1
Medicine® OBSERVATIONAL STUDY Association Between Vitamin D Receptor rs731236 (Taq1) Polymorphism and Risk for Restless Legs Syndrome in the Spanish Caucasian Population Fe´lix Javier Jime´nez-Jime´nez, MD, PhD, Elena Garcı´a-Martı´n, MD, PhD, Hortensia Alonso-Navarro, MD, PhD, Carmen Martı´nez, MD, PhD, Martı´n Zurdo, MD, Laura Turpı´n-Fenoll, MD, PhD, Jorge Milla´n-Pascual, MD, Teresa Adeva-Bartolome´, MD, PhD, Esther Cubo, MD, PhD, Francisco Navacerrada, MD, Ana Rojo-Sebastia´n, MD, Lluisa Rubio, MD, Sara Ortega-Cubero, MD, Pau Pastor, MD, PhD, Marisol Calleja, CN, Jose´ Francisco Plaza-Nieto, Bele´n Pilo-De-La-Fuente, MD, PhD, Margarita Arroyo-Solera, MD, Esteban Garcı´a-Albea, MD, PhD, and Jose´ A.G. Agu´ndez, MD, PhD Abstract: Several recent works suggest a possible role of vitamin D carrying the allelic variant rs731236G had an earlier age at onset, and deficiency in the etiology or restless legs syndrome (RLS). We analyzed those carrying the rs731236GG genotype had higher severity of RLS, the possible relationship of 2 common single nucleotide polymorphisms although these data disappeared after multivariate analyses. None of (SNPs) in the vitamin D3 receptor (VDR) gene with the risk for RLS. the SNPs studied was related with the positivity of family history of We studied the genotype and allelic variant frequencies of VDR RLS. rs2228570 and VDR rs731236 SNPs in 205 RLS patients and 445 These results suggest a modest, but significant association between healthy controls using a TaqMan essay. VDR rs731236 SNP and the risk for RLS. The frequencies of the rs731236AA genotype and the allelic variant (Medicine 94(47):e2125) rs731236A were significantly lower in RLS patients than in controls (P < 0.005 and < 0.01, respectively). -
Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia
Published OnlineFirst August 17, 2016; DOI: 10.1158/2159-8290.CD-16-0237 RESEARCH ARTICLE Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia Michael W.M. Kühn1,2, Evelyn Song1, Zhaohui Feng1, Amit Sinha1, Chun-Wei Chen1, Aniruddha J. Deshpande1, Monica Cusan1, Noushin Farnoud1, Annalisa Mupo3, Carolyn Grove4,5, Richard Koche1, James E. Bradner6, Elisa de Stanchina7, George S. Vassiliou3, Takayuki Hoshii1, and Scott A. Armstrong1,8 ABSTRACT Homeobox (HOX) proteins and the receptor tyrosine kinase FLT3 are frequently highly expressed and mutated in acute myeloid leukemia (AML). Aberrant HOX expression is found in nearly all AMLs that harbor a mutation in the Nucleophosmin (NPM1) gene, and FLT3 is concomitantly mutated in approximately 60% of these cases. Little is known about how mutant NPM1 (NPM1mut) cells maintain aberrant gene expression. Here, we demonstrate that the histone modi- fiers MLL1 and DOT1L controlHOX and FLT3 expression and differentiation in NPM1mut AML. Using a CRISPR/Cas9 genome editing domain screen, we show NPM1mut AML to be exceptionally dependent on the menin binding site in MLL1. Pharmacologic small-molecule inhibition of the menin–MLL1 protein interaction had profound antileukemic activity in human and murine models of NPM1mut AML. Combined pharmacologic inhibition of menin–MLL1 and DOT1L resulted in dramatic suppression of HOX and FLT3 expression, induction of differentiation, and superior activity against NPM1mut leukemia. SIGNIFICANCE: MLL1 and DOT1L are chromatin regulators that control HOX, MEIS1, and FLT3 expres- sion and are therapeutic targets in NPM1mut AML. Combinatorial small-molecule inhibition has synergistic on-target activity and constitutes a novel therapeutic concept for this common AML subtype. -

Supplementary Material Contents
Supplementary Material Contents Immune modulating proteins identified from exosomal samples.....................................................................2 Figure S1: Overlap between exosomal and soluble proteomes.................................................................................... 4 Bacterial strains:..............................................................................................................................................4 Figure S2: Variability between subjects of effects of exosomes on BL21-lux growth.................................................... 5 Figure S3: Early effects of exosomes on growth of BL21 E. coli .................................................................................... 5 Figure S4: Exosomal Lysis............................................................................................................................................ 6 Figure S5: Effect of pH on exosomal action.................................................................................................................. 7 Figure S6: Effect of exosomes on growth of UPEC (pH = 6.5) suspended in exosome-depleted urine supernatant ....... 8 Effective exosomal concentration....................................................................................................................8 Figure S7: Sample constitution for luminometry experiments..................................................................................... 8 Figure S8: Determining effective concentration .........................................................................................................