Isoschizomers.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Cloning of DNA Fragments Produced by Restriction
Proc. Natl. Acad. Sci. USA Vol. 73, No. 5, pp. 1537-1541, May 1976 Biochemistry Molecular cloning of DNA fragments produced by restriction endonucleases SailI and BamI (DNA joining/plasmid/insertional inactivation of genes/Drosophila melanogaster) DEAN H. HAMER AND CHARLES A. THOMAS, JR. Department of Biological Chemistry, Harvard Medical School, Boston, Massachusetts 02115 Communicated by Bernard D. Dats, February 25,1976 ABSTRACT The highly specific restriction endonucleases cleaves various DNAs about once every 8 kb (kilobases), as SalI and BamI produce DNA fragments with complementary, compared to about once every 4 kb for EcoRI (16, 18). The cohesive termini that can be covalently joined by DNA ligase. resulting fragments have cohesive termini, and can be joined The Escherichia coli kanamycin resistance factor pML21 has to one another in head-to-tail, head-to-head, and one Sall site, at which DNA can be inserted without interfering probably with the expression of drug resistance or replication of the tail-to-tail orientation. Another highly specific restriction en- plasmid. A more convenient cloning vehicle can be made with donuclease that produces cohesive termini is BamI, from Ba- the tetracycline resistance factor pSC101, since insertion of cillus amyloliqefaciens H (G. Wilson and F. Young, personal DNA either at its single site for Sall or at that for BamI inacti- communication). We have shown that it cleaves D. melano- vates plasmid-specified drug resistance but not replication. To gaster DNA about once every 6 kb. This report describes the take advantage of this insertional inactivation, pSC101 was construction and verification of plasmid vehicles that allow one joined to a ColEl-ampicillin resistance plasmid having no Sall site, and to a ColEl-kanamycin resistance plasmid having no to clone and amplify potentially any DNA fragment produced BamI site. -

Restriction Endonucleases
Molecular Biology Problem Solver: A Laboratory Guide. Edited by Alan S. Gerstein Copyright © 2001 by Wiley-Liss, Inc. ISBNs: 0-471-37972-7 (Paper); 0-471-22390-5 (Electronic) 9 Restriction Endonucleases Derek Robinson, Paul R. Walsh, and Joseph A. Bonventre Background Information . 226 Which Restriction Enzymes Are Commercially Available? . 226 Why Are Some Enzymes More Expensive Than Others? . 227 What Can You Do to Reduce the Cost of Working with Restriction Enzymes? . 228 If You Could Select among Several Restriction Enzymes for Your Application, What Criteria Should You Consider to Make the Most Appropriate Choice? . 229 What Are the General Properties of Restriction Endonucleases? . 232 What Insight Is Provided by a Restriction Enzyme’s Quality Control Data? . 233 How Stable Are Restriction Enzymes? . 236 How Stable Are Diluted Restriction Enzymes? . 236 Simple Digests . 236 How Should You Set up a Simple Restriction Digest? . 236 Is It Wise to Modify the Suggested Reaction Conditions? . 237 Complex Restriction Digestions . 239 How Can a Substrate Affect the Restriction Digest? . 239 Should You Alter the Reaction Volume and DNA Concentration? . 241 Double Digests: Simultaneous or Sequential? . 242 225 Genomic Digests . 244 When Preparing Genomic DNA for Southern Blotting, How Can You Determine If Complete Digestion Has Been Obtained? . 244 What Are Your Options If You Must Create Additional Rare or Unique Restriction Sites? . 247 Troubleshooting . 255 What Can Cause a Simple Restriction Digest to Fail? . 255 The Volume of Enzyme in the Vial Appears Very Low. Did Leakage Occur during Shipment? . 259 The Enzyme Shipment Sat on the Shipping Dock for Two Days. -

How to Clone Ref V1-2
A primer on cloning in E. coli Michael Nonet Department of Neuroscience Washington University Medical School St Louis, MO 63110 Draft 1.2.1 Nov. 10, 2019 Things to add: gel electrophoresis methods basic bacteriology methods Index Overview E. coli vectors! 6 Types of E. coli vectors! 6 Components of typical plasmid vectors! 6 Origin of replication! 6 Antibiotic resistance genes! 7 Master Cloning Sites! 7 LacZ"! 7 ccdB ! 8 Design of DNA constructs! 9 Choice of vector! 9 Source of insert! 9 Plasmids! 9 Genomic or cDNA! 9 Synthetic DNA! 10 Overview of different approaches to creating clones! 10 Restriction enzyme based cloning! 10 Single vs. double cut method! 11 Blunt vs “sticky” restriction sites! 11 Typical methodology for restriction enzyme cloning! 11 Golden Gate style cloning! 12 Typical methodology for Golden Gate cloning! 12 Gateway cloning! 13 Typical methodology for Gateway cloning! 14 Gibson assembly cloning! 15 Typical procedure for Gibson assembly! 15 Site directed mutagenesis! 16 PCR overlap approach! 16 DpnI mediated site directed mutagenesis! 16 Typical procedure for site directed mutagenesis! 17 Detailed methodology for all five methods Restriction enzyme cloning! 18 Designing the cloning strategy ! 18 Determining which vector to use! 18 Designing the insert fragment! 19 Designing primers! 19 Vector preparation! 19 Insert preparation! 19 Detailed protocol! 20 Step 1: Design oligonucleotides to amplify the product of interest! 20 Step 2: PCR amplification of insert DNA! 20 Step 3: Clean-up purification of PCR product! 21 Step 4: Digestion of products! 21 Step 5: Gel purification of products! 22 Step 6: Ligation! 22 Step 7: Transformation of E. -

The Cloning Guide the First Step Towards a Succesful Igem Project Igem Bonn NRP-UEA Igem
The Cloning Guide The first step towards a succesful iGEM Project iGEM Bonn NRP-UEA iGEM iGEM Manchester-Graz iGEM Evry iGEM Vanderbilt iGEM Minnesota Carnegie Mellon iGEM iGEM Sydney UIUC iGEM iGEM York iGEM Toulouse iGEM Pasteur UCLA iGEM iGEM Stockholm Preface The cloning guide which is lying before you or on your desktop is a document brought to you by iGEM TU Eindhoven in collaboration with numerous iGEM (International Genetically Engi- neered Machine) teams during iGEM 2015. An important part in the iGEM competition is the collabration of your own team with other teams. These collaborations are fully in line with the dedications of the iGEM Foundation on education and competition, the advancement of syn- thetic biology, and the development of an open community and collaboration. Collaborations between groups of (undergrad and overgrad) students can lead to nice products, as we have tried to provide one for you in 2015. In order to compile a cloning guide, several iGEM teams from all over the world have been con- tacted to cooperate on this. Finally fifiteen teams contributed in a fantastic way and a cloning guide consisting of the basics about nine different cloning methods and experiences of teams working with them has been realized. Without the help of all collaborating teams this guide could never have been realized, so we will thank all teams in advance. This guide may be of great help when new iGEM teams (edition 2016 and later) are at the point of designing their project. How to assemble the construct for you project, is an important choice which is possibly somewhat easier to make after reading this guide. -

Bpuami: a Novel Saci Neoschizomer from Bacillus Pumilus Discovered in an Isolate from Amazon Basin, Recognizing 5'-Gag↓Ctc-3'
Brazilian Journal of Microbiology (2006) 37:96-100 ISSN 1517-8382 BPUAMI: A NOVEL SACI NEOSCHIZOMER FROM BACILLUS PUMILUS DISCOVERED IN AN ISOLATE FROM AMAZON BASIN, RECOGNIZING 5'-GAG↓CTC-3' Jocelei M. Chies1,2,*; Ana C. de O. Dias1; Hélio M. M. Maia3; Spartaco Astolfi-Filho2,3 1Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul, RS, Brasil; 2Curso de Pós-Graduação em Biologia Molecular, Universidade de Brasília, Brasília, DF, Brasil; 3Centro de Apoio Multidisciplinar, Universidade Federal do Amazonas, Manaus, AM, Brasil Submitted: June 27, 2005; Returned to authors for corrections: November 16, 2005; Approved: January 12, 2006 ABSTRACT A strain of Bacillus pumilus was isolated and identified from water samples collected from a small affluent of the Amazon River. Type II restriction endonuclease activity was detected in these bacteria. The enzyme was purified and the molecular weight of the native protein estimated by gel filtration and SDS-PAGE. The optimum pH, temperature and salt requirements were determined. Quality control assays showed the complete absence of “nonspecific nucleases.” Restriction cleavage analysis and DNA sequencing of restriction fragments allowed the unequivocal demonstration of 5´GAG↓CTC3´ as the recognition sequence. This enzyme was named BpuAmI and is apparently a neoschizomer of the prototype restriction endonuclease SacI. This is the first report of an isoschizomer and/or neoschizomer of the prototype SacI identified in the genus Bacillus. Key words: type II restriction endonuclease, BpuAmI, SacI, neoschizomers, Bacillus pumilus INTRODUCTION thousands of taxonomically diverse bacteria for enzymes with new characteristics. However, to our knowledge, there has Restriction endonucleases are enzymes which recognize not been any report to date of an isoschizomer and/or short DNA sequences and cleave DNA in both strands. -

1. Restriction Endonucleases (® Fermentas 2006)
We guarantee that these products are Introduction ..........................................................................................................2 free of contaminating activities. Restriction Endonucleases Guide Our stringent quality control with the most FastDigest™ Restriction Endonucleases .........................................................2 Fermentas Restriction Endonucleases............................................................3 advanced tests guarantees you pure 1 Alphabetic List of Commercially Available Restriction Endonucleases.........6 products for your experiments. ISO9001 Recognition Specifi cities...............................................................................16 and ISO14001 is your assurance of con- Commercial Restriction Enzymes Sorted by the Type of DNA Ends Generated sistency and lot-to-lot reproducibility. Enzymes Generating 5’-protruding Ends.......................................................18 Enzymes Generating 3’-protruding Ends.......................................................18 ™ PureExtreme Quality will provide the Enzymes Generating Blunt Ends...................................................................19 performance you need for your most Enzymes Cleaving DNA on the Both Sides of Their Recognition Sequences.....19 demanding experiments. PureExtreme™ Quality PureExtreme™ Quality Guarantee..................................................................20 Activity Assay.................................................................................................20 -

A Short History of the Restriction Enzymes Wil A
Published online 18 October 2013 Nucleic Acids Research, 2014, Vol. 42, No. 1 3–19 doi:10.1093/nar/gkt990 NAR Breakthrough Article SURVEY AND SUMMARY Highlights of the DNA cutters: a short history of the restriction enzymes Wil A. M. Loenen1,*, David T. F. Dryden2,*, Elisabeth A. Raleigh3,*, Geoffrey G. Wilson3,* and Noreen E. Murrayy 1Leiden University Medical Center, Leiden, the Netherlands, 2EaStChemSchool of Chemistry, University of Edinburgh, West Mains Road, Edinburgh EH9 3JJ, Scotland, UK and 3New England Biolabs, Inc., 240 County Road, Ipswich, MA 01938, USA Received August 14, 2013; Revised September 24, 2013; Accepted October 2, 2013 ABSTRACT Type II REases represent the largest group of characterized enzymes owing to their usefulness as tools for recombinant In the early 1950’s, ‘host-controlled variation in DNA technology, and they have been studied extensively. bacterial viruses’ was reported as a non-hereditary Over 300 Type II REases, with >200 different sequence- phenomenon: one cycle of viral growth on certain specificities, are commercially available. Far fewer Type I, bacterial hosts affected the ability of progeny virus III and IV enzymes have been characterized, but putative to grow on other hosts by either restricting or examples are being identified daily through bioinformatic enlarging their host range. Unlike mutation, this analysis of sequenced genomes (Table 1). change was reversible, and one cycle of growth in Here we present a non-specialists perspective on import- the previous host returned the virus to its original ant events in the discovery and understanding of REases. form. These simple observations heralded the dis- Studies of these enzymes have generated a wealth of covery of the endonuclease and methyltransferase information regarding DNA–protein interactions and catalysis, protein family relationships, control of restric- activities of what are now termed Type I, II, III and tion activity and plasticity of protein domains, as well IV DNA restriction-modification systems. -
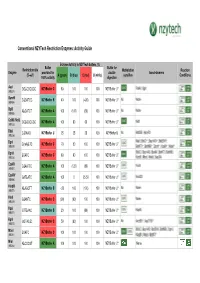
Conventional Nzytech Restriction Enzymes: Activity Guide
Conventional NZYTech Restriction Enzymes: Activity Guide Enzyme Activity in NZYTech buffers (%) Buffer Buffer for Restriction site Methylation Reaction Enzyme provided for double Isoschizomers (5’ →→→3’) A (green) B (blue) C (red) U (white) sensitive Conditions 100% activity digestion AscI GG ↓↓↓CGCGCC NZYBuffer C 80 100 100 100 NZYBuffer U * PalAI, SgsI (MB231) BamHI G↓↓↓GATCC NZYBuffer B 40 100 (<20) 100 NZYBuffer U * No None (MB064) BglII A↓↓↓GATCT NZYBuffer A 100 (100) (60) 100 NZYBuffer U * No None (MB065) CciNI (NotI) GC ↓↓↓GGCCGC NZYBuffer A 100 80 60 100 NZYBuffer U * NotI (MB153) DdeI C↓↓↓TNAG NZYBuffer U 25 25 25 100 NZYBuffer U No BstDEI, HpyF3I (MB236) MalI, BfuCI ♦♦♦, Bsp143I ♦♦♦, BstENII ♦♦♦, DpnI G(mA)↓↓↓TC NZYBuffer C 70 50 100 100 NZYBuffer U * (MB078) BstKTI ♦♦♦, BstMBI ♦♦♦, DpnII ♦♦♦, Kzo9I ♦♦♦, NdeII ♦♦♦ BfuCI, Bsp143I, BssMI, BstKTI, BstMBI, DpnII ↓↓↓GATC NZYBuffer C 80 80 100 100 NZYBuffer U * (MB233) Kzo9I, MboI, NdeII, Sau3AI EcoRI G↓↓↓AATTC NZYBuffer A 100 (120) (80) 100 NZYBuffer U * FunII (MB067) EcoRV GAT ↓↓↓ATC NZYBuffer A 100 0 25-50 100 NZYBuffer U * Eco32I (MB068) HindIII A↓↓↓AGCTT NZYBuffer B <20 100 (100) 100 NZYBuffer U * No None (MB070) HinfI G↓↓↓ANTC NZYBuffer C (90) (90) 100 100 NZYBuffer U * None (MB239) HpaI GTT ↓↓↓AAC NZYBuffer B 20 100 (80) 100 NZYBuffer U * KspAI (MB071) KpnI GGTAC ↓↓↓C NZYBuffer C 50 (80) 100 100 NZYBuffer U * No Acc65I ♦♦♦, Asp718I ♦♦♦ (MB072) BfuCI, Bsp143I, BstENII, BstMBI, DpnII, MboI ↓↓↓GATC NZYBuffer C 100 100 100 100 NZYBuffer U * (MB241) Kzo9II, NdeII, BstKTI ♦♦♦ MluI -

Promiscuous Restriction Is a Cellular Defense Strategy That Confers Fitness
Promiscuous restriction is a cellular defense strategy PNAS PLUS that confers fitness advantage to bacteria Kommireddy Vasua, Easa Nagamalleswaria, and Valakunja Nagarajaa,b,1 aDepartment of Microbiology and Cell Biology, Indian Institute of Science, Bangalore 560012, India; and bJawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560064, India Edited by Werner Arber, der Universitat Basel, Basel, Switzerland, and approved March 20, 2012 (received for review November 22, 2011) Most bacterial genomes harbor restriction–modification systems, ognition and cofactor utilization, whereas the cognate MTase is encoding a REase and its cognate MTase. On attack by a foreign very site-specific (14). In addition to its recognition sequence, DNA, the REase recognizes it as nonself and subjects it to restric- GGTACC, the enzyme binds and cleaves a number of noncanoni- tion. Should REases be highly specific for targeting the invading cal sequences (i.e., TGTACC, GTTACC, GATACC, GGAACC, foreign DNA? It is often considered to be the case. However, when GGTCCC, GGTATC, GGTACG, GGTACT), with the binding bacteria harboring a promiscuous or high-fidelity variant of the affinity ranging from 8 to 35 nM (14, 15). Accordingly, many of fi REase were challenged with bacteriophages, tness was maximal these sites are cleaved efficiently with kcat values varying from 0.06 − − under conditions of catalytic promiscuity. We also delineate pos- to 0.15 min 1 vs. 4.3 min 1 for canonical sites (15). The promis- sible mechanisms by which the REase recognizes the chromosome cuous activity of the enzyme is directed by a number of cofactors. as self at the noncanonical sites, thereby preventing lethal dsDNA In the presence of the physiological levels of Mg2+, the most breaks. -

Biology of DNA Restriction THOMAS A
MICROBIOLOGICAL REVIEWS, June 1993, p. 434-450 Vol. 57, No. 2 0146-0749/93/020434-17$02.00/0 Copyright © 1993, American Society for Microbiology Biology of DNA Restriction THOMAS A. BICKLE'* AND DETLEV H. KRUGER2 Department ofMicrobiology, Biozentrum, Basel University, Klingelbergstrasse 70, CH-4056 Basel, Switzerland, 1 and Institute of Virology, Charite School ofMedicine, Humboldt University, D-0-1040 Berlin, Gernany2 INTRODUCTION ........................................................................ 434 TYPE I R-M SYSTEMS ........................................................................ 434 Type I Systems Form Families of Related Enzymes ...................................................................435 Structure of hsd Genes ........................................................................ 435 Evolution of DNA Sequence Recognition by Recombination between hsdS Genes .........................***436 Mutations Affecting Modification Activity........................................................................ 437 TYPE II R-M SYSTEMS........................................................................ 437 Evolutionary Aspects ........................................................................ 437 Control of Expression of Type II RM Genes .....................................................438 Cytosine Can Be Methylated on Either C-5 Or NA: Consequences for Mutagenesis...............438 Type II Restriction Endonucleases That Require Two Recognition Sites for Cleavage.439 What Is the Function of Type IIS Enzymes.440 -

Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS PERSONAL
Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS New England Biolabs 240 County Road, Ipswich, MA 02138 USA Email: [email protected] Telephone: (978) 380-7405 / Fax: (978) 380-7406 PERSONAL Born on September 6, 1943, Derby, England EDUCATION 1962-1965 University of Sheffield, Sheffield, England B.Sc. in Chemistry 1966-1968 University of Sheffield, Sheffield, England Ph.D. in Organic Chemistry POSITIONS 2005- Chief Scientific Officer, New England Biolabs 1992-2005 Research Director, New England Biolabs 1986-92 Assistant Director for Research, Cold Spring Harbor Laboratory 1972-86 Senior Staff Investigator, Cold Spring Harbor Laboratory 1971-1972 Research Associate in Biochemistry, Harvard University 1969-1970 Research Fellow, Harvard University OUTSIDE ACTIVITIES 1974-1992 Consultant and Chairman of Scientific Advisory Board New England Biolabs 1977-1985 Scientific Advisory Board, Genex Corp. 1977-1987 Editorial Board: Nucleic Acids Research 1979-1984 Editorial Board: Journal of Biological Chemistry 1982-1989 Member: National Advisory Committee of GENBANK 1984-1986 Member: National Advisory Committee of BIONET 1985-1988 Panel member: NIH Study Section in Biochemistry. 1985-2002 Editorial Board: Bioinformatics (formerly CABIOS) 1987-1990 Chairman: National Advisory Committee of BIONET 1987-2009 Senior Executive Editor: Nucleic Acids Research 1990-1992 Panel member: NCI Cancer Centers Support Grant Review Committee 1993-1995 Panel member: NLM Study Section/Comp. Biol. 1994-2000 Scientific Advisory Board, Molecular Tool 1994- Patron of the Oxford International Biomedical Center 1996-1998 Visiting Professor, University of Bath, UK. 1996-2000 Chairman, NCI Board of Scientific Counselors 1996-1999 Scientific Advisory Board, Oxford Molecular Group 1997-2001 Editorial Board: Current Opinion Chem. Biol. -

NEB Golden Gate Assembly Kit (Bsai-Hfv2) E1601 Manual
INSTRUCTION MANUAL NEB® Golden Gate Assembly Kit (BsaI-HF®v2) NEB #E1601S/L 20/100 reactions Version 2.0_2/20 Table of Contents Golden Gate Assembly Overview ............................................................................................................................................................... 2 pGGAselect Destination Plasmid Description ............................................................................................................................................ 3 Insert Considerations ................................................................................................................................................................................... 4 Golden Gate Assembly Transformation and Plating Protocols ................................................................................................................... 5 Screening Protocols ..................................................................................................................................................................................... 6 Frequently Asked Questions (FAQs) .......................................................................................................................................................... 6 Golden Gate Assembly Tips ........................................................................................................................................................................ 8 Specifications ..............................................................................................................................................................................................