Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS PERSONAL
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Extensive Experience. Superior Quality
EXTENSIVE EXPERIENCE. SUPERIOR QUALITY. INNOVATIVE SOLUTIONS. R&D AND MANUFACTURING SUPPORT FOR THE BIOTECHNOLOGY AND BIOPHARMA INDUSTRY LET NEB’S SCIENTIFIC EXPERTISE HELP DRIVE YOUR INNOVATIONS FORWARD In recent years, there has been a dramatic rise in the number of biologics and companion diagnostics developed and commercialized, and these important areas of biotechnology are more reliant than ever on cutting edge molecular biology tools and techniques. For instance, companion diagnostics, based on DNA amplification, can foster a better match of effective therapies with patients; personalized medicines, utilizing next-generation sequencing, can also help tailor treatments for specific patients; and a wide range of biologics is now being developed as a result of advancements in recombinant technologies. 2 COLLABORATION We’re here to champion your efforts. We have extensive scientific expertise and can devote the necessary attention to developing solutions for your specific needs – along with the manufacturing capacity to scale up quickly. Whether you’re looking for a custom version of an existing product, to find a new way around a development roadblock, or to license one of our technologies, our team is ready to work with you. PRODUCT PORTFOLIO We can provide an entire suite of enzymes THE NEB DIFFERENCE that have the potential to accelerate your discovery efforts. Our expertise in At NEB, we have decades of experience enzymology has enabled us to develop unique enzymes that enable faster, more robust in practicing molecular biology, which has workflows. Further, enzymes can be provided both in small aliquots and in bulk, in different led to the introduction of a broad product formats (liquid, lyophilized and glycerol-free), portfolio that has the potential to touch as well as packaged into complete kits. -

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -

“ ” Enabling Real-Time Business Growth with SAP S/4HANA® In
Enabling real-time business growth with SAP S/4HANA® in the cloud Case Study: New England Biolabs An SAP HANA® and S/4 Migration Case Study Scientists today are constantly under pressure to quickly already difficult task, consistent, double-digit growth was understand the genetic makeup of new bacteria strains, rapidly powering the business forward, but exposed gaps in help diagnose new diseases and create life-saving the IT infrastructure. treatments for use worldwide. Their implementation of the SAP ECC 6.0 platform was As a global leader in the discovery, development and lacking key functionality in the areas of production planning, commercialization of recombinant and native enzymes for eProcurement and product costing. The need for a stable, genomic research, New England Biolabs (NEB) is an and more importantly scalable platform to support the innovator of technologies from basic molecular biology rapidly evolving business became critical. As NEB began to through to sample preparation for next-generation evaluate their options, they were faced with the realities of sequencing and high throughput genomics for the life having a small IT team who didn’t have the necessary SAP sciences industry. Headquartered in the US with seven technical experience to implement a “cloud first” strategy global subsidiaries, they are at the forefront of medical and and support a large scale ERP effort. technological developments. The Competitive Edge Assessing the Current Landscape NEB began the process of looking for the right provider to A key offering of New England Biolabs business is the augment their team and initiatives. “We put out a bid to NEBnow Freezer Program, which consists of teams working see which hosting provider best aligned with our needs, and closely with institutions to customize inventory best suited Symmetry’s full suite of services stood out above the rest,” to research programs. -

Catalogue of Bacteria Shapes
We first tried to use the most general shape associated with each genus, which are often consistent across species (spp.) (first choice for shape). If there was documented species variability, either the most common species (second choice for shape) or well known species (third choice for shape) is shown. Corynebacterium: pleomorphic bacilli. Due to their snapping type of division, cells often lie in clusters resembling chinese letters (https://microbewiki.kenyon.edu/index.php/Corynebacterium) Shown is Corynebacterium diphtheriae Figure 1. Stained Corynebacterium cells. The "barred" appearance is due to the presence of polyphosphate inclusions called metachromatic granules. Note also the characteristic "Chinese-letter" arrangement of cells. (http:// textbookofbacteriology.net/diphtheria.html) Lactobacillus: Lactobacilli are rod-shaped, Gram-positive, fermentative, organotrophs. They are usually straight, although they can form spiral or coccobacillary forms under certain conditions. (https://microbewiki.kenyon.edu/index.php/ Lactobacillus) Porphyromonas: A genus of small anaerobic gram-negative nonmotile cocci and usually short rods thatproduce smooth, gray to black pigmented colonies the size of which varies with the species. (http:// medical-dictionary.thefreedictionary.com/Porphyromonas) Shown: Porphyromonas gingivalis Moraxella: Moraxella is a genus of Gram-negative bacteria in the Moraxellaceae family. It is named after the Swiss ophthalmologist Victor Morax. The organisms are short rods, coccobacilli or, as in the case of Moraxella catarrhalis, diplococci in morphology (https://en.wikipedia.org/wiki/Moraxella). *This one could be changed to a diplococcus shape because of moraxella catarrhalis, but i think the short rods are fair given the number of other moraxella with them. Jeotgalicoccus: Jeotgalicoccus is a genus of Gram-positive, facultatively anaerobic, and halotolerant to halophilicbacteria. -

CALIFORNIA STATE UNIVERSITY, NORTHRIDGE Comparative
CALIFORNIA STATE UNIVERSITY, NORTHRIDGE Comparative Genomics and Epigenomics of Sporosarcina ureae A thesis submitted in partial fulfillment of the requirement for the degree of Master of Science in Biology By Andrew Oliver August 2016 The thesis of Andrew Oliver is approved by: _________________________________________ ____________ Sean Murray, Ph.D. Date _________________________________________ ____________ Gilberto Flores, Ph.D. Date _________________________________________ ____________ Kerry Cooper, Ph.D., Chair Date California State University, Northridge ii Acknowledgments First and foremost, a special thanks to my advisor, Dr. Kerry Cooper, for his advice and, above all, his patience. If I can be half the scientist you are someday, I would be thrilled. I would like to also thank everyone in the Cooper lab, especially my colleagues Courtney Sams and Tabitha Bayangnos. It was a privilege to work along side you. More thanks to my committee members, Dr. Gilberto Flores and Dr. Sean Murray. Dr. Flores, you were instrumental in guiding me to ask the right questions regarding bacterial taxonomy. Dr. Murray, your contributions to my graduate studies would make this section run on for pages. I thank you for taking me under your wing from the beginning. Acknowledgement and thanks to the Baresi lab, especially Dr. Larry Baresi and Tania Kurbessoian for their partnership in this research. Also to Bernardine Pregerson for all the work that lays at the foundation of this study. This research would not be what it is without the help of my childhood friend, Matthew Kay. You wrote programs, taught me coding languages, and challenged me to go digging for answers to very difficult questions. -

Product Sheet Info
Product Information Sheet for HM-331 Sporosarcina sp., Strain 2681 Incubation: Temperature: 37°C Atmosphere: Aerobic Catalog No. HM-331 Propagation: 1. Keep vial frozen until ready for use, then thaw. For research use only. Not for human use. 2. Transfer the entire thawed aliquot into a single tube of broth. Contributor: 3. Use several drops of the suspension to inoculate an Kimberlee A. Musser, Ph.D., Chief, Bacterial Diseases, agar slant and/or plate. Division of Infectious Diseases, Wadsworth Center, New York 4. Incubate the tubes and plate at 37°C for 48 hours. State Department of Health, Albany, New York Citation: Manufacturer: NIH Biodefense and Emerging Infections Acknowledgment for publications should read “The following reagent was obtained through the NIH Biodefense and Research Resource Repository Emerging Infections Research Resources Repository, NIAID, NIH as part of the Human Microbiome Project: Sporosarcina Product Description: sp., Strain 2681, HM-331.” Bacteria Classification: Planococcaceae, Sporosarcina Species: Sporosarcina sp. Biosafety Level: 2 Strain: 2681 Original Source: Sporosarcina sp., strain 2681 was isolated Appropriate safety procedures should always be used with 1 this material. Laboratory safety is discussed in the following from human blood. Comments: Sporosarcina sp., strain 2681 is a reference publication: U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and genome for The Human Microbiome Project (HMP). HMP Prevention, and National Institutes of Health. Biosafety in is an initiative to identify and characterize human microbial flora. The complete genome of Sporosarcina sp., strain Microbiological and Biomedical Laboratories. 5th ed. Washington, DC: U.S. Government Printing Office, 2007; see 2681 is currently being sequenced at the Human Genome www.cdc.gov/od/ohs/biosfty/bmbl5/bmbl5toc.htm. -

Bpuami: a Novel Saci Neoschizomer from Bacillus Pumilus Discovered in an Isolate from Amazon Basin, Recognizing 5'-Gag↓Ctc-3'
Brazilian Journal of Microbiology (2006) 37:96-100 ISSN 1517-8382 BPUAMI: A NOVEL SACI NEOSCHIZOMER FROM BACILLUS PUMILUS DISCOVERED IN AN ISOLATE FROM AMAZON BASIN, RECOGNIZING 5'-GAG↓CTC-3' Jocelei M. Chies1,2,*; Ana C. de O. Dias1; Hélio M. M. Maia3; Spartaco Astolfi-Filho2,3 1Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul, RS, Brasil; 2Curso de Pós-Graduação em Biologia Molecular, Universidade de Brasília, Brasília, DF, Brasil; 3Centro de Apoio Multidisciplinar, Universidade Federal do Amazonas, Manaus, AM, Brasil Submitted: June 27, 2005; Returned to authors for corrections: November 16, 2005; Approved: January 12, 2006 ABSTRACT A strain of Bacillus pumilus was isolated and identified from water samples collected from a small affluent of the Amazon River. Type II restriction endonuclease activity was detected in these bacteria. The enzyme was purified and the molecular weight of the native protein estimated by gel filtration and SDS-PAGE. The optimum pH, temperature and salt requirements were determined. Quality control assays showed the complete absence of “nonspecific nucleases.” Restriction cleavage analysis and DNA sequencing of restriction fragments allowed the unequivocal demonstration of 5´GAG↓CTC3´ as the recognition sequence. This enzyme was named BpuAmI and is apparently a neoschizomer of the prototype restriction endonuclease SacI. This is the first report of an isoschizomer and/or neoschizomer of the prototype SacI identified in the genus Bacillus. Key words: type II restriction endonuclease, BpuAmI, SacI, neoschizomers, Bacillus pumilus INTRODUCTION thousands of taxonomically diverse bacteria for enzymes with new characteristics. However, to our knowledge, there has Restriction endonucleases are enzymes which recognize not been any report to date of an isoschizomer and/or short DNA sequences and cleave DNA in both strands. -

Research Journal of Pharmaceutical, Biological and Chemical Sciences
ISSN: 0975-8585 Research Journal of Pharmaceutical, Biological and Chemical Sciences Florula of Larval and Imaginal Phases of the Volfartova Fly (Wohlfarthia magnifica) In the Conditions of the Steppe Zone of The Pavlodar Region. A A Bitkeyeva1* and L T Bulekbayeva2. 1Senior teacher, Master of Ecology, Pavlodar State University named after S. Toraygyrov, The Republic of Kazakhstan. 2Associate professor, Candidate of Biological Sciences, Pavlodar State Pedagogical Institute, Republic of Kazakhstan. ABSTRACT Groups of bacteria were found during research in a steppe zone of the Pavlodar region, belonging to 3 families: Baccilaceae, Micrococcaceae, Enterobacteriacea. 13 species of pathogenic and opportunistic bacteria are obtained and identified, which cause diseases. Reception of agents from flies of Wohlfartia magnifica family in region farms forces to pay attention to quite real possibility and contagion of various infections. It creates the menacing epidemiological and epizootiology situation on the adjacent to farms of populated places, as flies with excrements can infect forages and migrate on considerable distances. Keywords: bacteria, diseases, infections, larvaes, microorganisms, flies, sheep, pathogenic microorganisms, carriers. *Corresponding author July– August 2015 RJPBCS 6(4) Page No. 2069 ISSN: 0975-8585 INTRODUCTION Flies are known as carriers of causative agents of dangerous infectious and invasive diseases. Therefore, in the populated places and on the pastures, studying of microbal and helminthosis impurity of flies represents scientific and practical interest. Epidemiological value of flies was opened by E.N. Pavlovskiy and V.P. Derbeneva-Ukhova, they participate in distribution about 70 pathogenic microflora, and including agents of a tularemia, anthrax, diphtheria, cholera, plague, a crab hand, etc. [2; 8; 12]. -
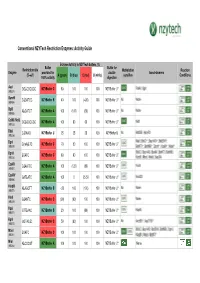
Conventional Nzytech Restriction Enzymes: Activity Guide
Conventional NZYTech Restriction Enzymes: Activity Guide Enzyme Activity in NZYTech buffers (%) Buffer Buffer for Restriction site Methylation Reaction Enzyme provided for double Isoschizomers (5’ →→→3’) A (green) B (blue) C (red) U (white) sensitive Conditions 100% activity digestion AscI GG ↓↓↓CGCGCC NZYBuffer C 80 100 100 100 NZYBuffer U * PalAI, SgsI (MB231) BamHI G↓↓↓GATCC NZYBuffer B 40 100 (<20) 100 NZYBuffer U * No None (MB064) BglII A↓↓↓GATCT NZYBuffer A 100 (100) (60) 100 NZYBuffer U * No None (MB065) CciNI (NotI) GC ↓↓↓GGCCGC NZYBuffer A 100 80 60 100 NZYBuffer U * NotI (MB153) DdeI C↓↓↓TNAG NZYBuffer U 25 25 25 100 NZYBuffer U No BstDEI, HpyF3I (MB236) MalI, BfuCI ♦♦♦, Bsp143I ♦♦♦, BstENII ♦♦♦, DpnI G(mA)↓↓↓TC NZYBuffer C 70 50 100 100 NZYBuffer U * (MB078) BstKTI ♦♦♦, BstMBI ♦♦♦, DpnII ♦♦♦, Kzo9I ♦♦♦, NdeII ♦♦♦ BfuCI, Bsp143I, BssMI, BstKTI, BstMBI, DpnII ↓↓↓GATC NZYBuffer C 80 80 100 100 NZYBuffer U * (MB233) Kzo9I, MboI, NdeII, Sau3AI EcoRI G↓↓↓AATTC NZYBuffer A 100 (120) (80) 100 NZYBuffer U * FunII (MB067) EcoRV GAT ↓↓↓ATC NZYBuffer A 100 0 25-50 100 NZYBuffer U * Eco32I (MB068) HindIII A↓↓↓AGCTT NZYBuffer B <20 100 (100) 100 NZYBuffer U * No None (MB070) HinfI G↓↓↓ANTC NZYBuffer C (90) (90) 100 100 NZYBuffer U * None (MB239) HpaI GTT ↓↓↓AAC NZYBuffer B 20 100 (80) 100 NZYBuffer U * KspAI (MB071) KpnI GGTAC ↓↓↓C NZYBuffer C 50 (80) 100 100 NZYBuffer U * No Acc65I ♦♦♦, Asp718I ♦♦♦ (MB072) BfuCI, Bsp143I, BstENII, BstMBI, DpnII, MboI ↓↓↓GATC NZYBuffer C 100 100 100 100 NZYBuffer U * (MB241) Kzo9II, NdeII, BstKTI ♦♦♦ MluI -

DNA Methylation Patterns and Cancer
restriction/modification system, which brought Werner Arber, Daniel Nathans and Hamilton Smith the 1978 Nobel Prize in Physiology or Medicine, and made restriction enzymes the primary tools of Charles Rodolphe Brupbacher Foundation molecular biology. Four decades have passed since then, but the role of 5-methylcytosine in eukaryotic DNA metabolism is still shrouded in mystery. We know that the sperm methylation pattern is largely The erased after fertilization and that methylation is gradually reintroduced Charles Rodolphe Brupbacher Prize during embryogenesis and differentiation, but the processes that for Cancer Research 2017 regulate the cell type- and tissue-specific methylation patterns remain is awarded to to be elucidated. We have also learned that DNA can be not only methylated, but also demethylated, and that aberrant methylation can lead to disease - including cancer. Again, how these processes are regulated remains to be discovered. However, we have learnt a great Sir Adrian Peter Bird, deal about 5-methylcytosine metabolism during the past three decades and much of our knowledge came from the laboratory of Adrian Bird. PhD Adrian spent his doctoral and postdoctoral time in Max Birnstiel’s for his contributions to our understanding laboratory, first in Edinburgh and then in Zurich, studying the amplification of ribosomal DNA in Xenopus laevis. In this organism, of the role of DNA methylation in genomic rDNA in somatic tissues is highly methylated, while the development and disease extrachromosomal amplicons are unmethylated. When he returned to Edinburgh to establish his own group, Adrian set out to study The President The President of the Foundation of the Scientific Advisory Board the methylation pattern of these loci using the newly-available methylation-sensitive restriction enzymes. -

Restriction Enzymes Are Molecular Scissors
Molecular Scissors Restriction enzymes are molecular scissors DR. ABUL HASAN SARDAR Assistant Professor and Head Department of Microbiology Sarsuna College RESTRICTION ENZYMES • A restriction enzyme (or restriction endonuclease) is an enzyme that cuts double- stranded or single stranded DNA at specific recognition nucleotide sequences known as restriction sites. Property of restriction enzymes • They that link adjacent nucleotides in DNA molecules. HOW RESTRICTION ENZYMES WORKS? • Restriction enzymes recognize a specific sequence of nucleotides, and produce a double-stranded cut in the DNA, these cuts are of two types: • BLUNT ENDS. • STICKY ENDS. Blunt end Sticky end BLUNT ENDS • These blunt ended fragments can be joined to any other DNA fragment with blunt ends. • Enzymes useful for certain types of DNA cloning experiments “STICKY ENDS” ARE USEFUL DNA fragments with complimentary sticky ends can be combined to create new molecules which allows the creation and manipulation of DNA sequences from different sources. • While recognition sequences vary widely , with lengths between 4 and 8 nucleotides, many of them are palindromic. PALINDROMES IN DNA SEQUENCES Genetic palindromes are similar to verbal palindromes. A palindromic sequence in DNA is one in which the 5’ to 3’ base pair sequence is identical on both strands (the 5’ and 3’ ends refers to the chemical structure of the DNA). PALINDROME SEQUENCES • The mirror like palindrome in which the same forward and backwards are on a single strand of DNA strand, as in GTAATG • The Inverted repeat palindromes is also a sequence that reads the same forward and backwards, but the forward and backward sequences are found in complementary DNA strands (GTATAC being complementary to CATATG) • Inverted repeat palindromes are more common and have greater biological importance than mirror- like palindromes. -

Biography of Invited Speakers
Biography of Invited Speakers Alan Ferst Alan Fersht is a group leader at the MRC Laboratory of Molecular Biology. He enjoys combining methods and ideas of molecular and structural biology with those from biophysics and chemistry to study the structure, activity, stability and folding of proteins, and the role of protein misfolding and instability in cancer and disease. His recent previous positions have been Herchel Smith Professor of Organic Chemistry at Cambridge University and Director of the MRC Centre for Protein Engineering. Currently, his major work is using structural and biophysical methods to study how mutation affects proteins in the cell cycle, particularly the tumour suppressor p53, in order to design novel anti‐cancer drugs that function by restoring the activity of mutated proteins. He is solving the structures of p53 and its negative regulator Mdm2, which are paradigms for partly intrinsically disordered proteins, by combining a variety of structural methods. Alan Fersht is a Fellow of the Royal Society, Foreign Associate of the National Academy of Sciences (USA), Honorary Foreign Member of the American Academy of Arts and Sciences, Member of EMBO and Member of Academia Europaea. He has won several international awards, including: the FEBS Anniversary Prize, 1980; Novo Biotechnology Award, 1986; Charmian Medal of the Royal Society of Chemistry, 1986 (for Enzymology); The Gabor Medal of the Royal Society, 1991 (for Molecular Biology); Max Tishler Lecture and Prize, Harvard University, 1992; FEBS Datta Lecture and Medal,