A Novel Pseudomonas Geniculata AGE Family Epimerase/Isomerase and Its Application in D-Mannose Synthesis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Orlistat, a Novel Potent Antitumor Agent for Ovarian Cancer: Proteomic Analysis of Ovarian Cancer Cells Treated with Orlistat
INTERNATIONAL JOURNAL OF ONCOLOGY 41: 523-532, 2012 Orlistat, a novel potent antitumor agent for ovarian cancer: proteomic analysis of ovarian cancer cells treated with Orlistat HUI-QIONG HUANG1*, JING TANG1*, SHENG-TAO ZHOU1, TAO YI1, HONG-LING PENG1, GUO-BO SHEN2, NA XIE2, KAI HUANG2, TAO YANG2, JIN-HUA WU2, CAN-HUA HUANG2, YU-QUAN WEI2 and XIA ZHAO1,2 1Gynecological Oncology of Biotherapy Laboratory, Department of Gynecology and Obstetrics, West China Second Hospital, Sichuan University, Chengdu, Sichuan; 2State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, Sichuan, P.R. China Received February 9, 2012; Accepted March 19, 2012 DOI: 10.3892/ijo.2012.1465 Abstract. Orlistat is an orally administered anti-obesity drug larly PKM2. These changes confirmed our hypothesis that that has shown significant antitumor activity in a variety of Orlistat is a potential inhibitor of ovarian cancer and can be tumor cells. To identify the proteins involved in its antitumor used as a novel adjuvant antitumor agent. activity, we employed a proteomic approach to reveal protein expression changes in the human ovarian cancer cell line Introduction SKOV3, following Orlistat treatment. Protein expression profiles were analyzed by 2-dimensional polyacrylamide In the 1920s, the Nobel Prize winner Otto Warburg observed gel electrophoresis (2-DE) and protein identification was a marked increase in glycolysis and enhanced lactate produc- performed on a MALDI-Q-TOF MS/MS instrument. More tion in tumor cells even when maintained in conditions of high than 110 differentially expressed proteins were visualized oxygen tension (termed Warburg effect), leading to widespread by 2-DE and Coomassie brilliant blue staining. -

Masterarbeit / Master's Thesis
MASTERARBEIT / MASTER’S THESIS Titel der Masterarbeit / Title of the Master‘s Thesis „Physiology, Metabolism and Biohydrogen Production of Desulfurococcus fermentans“ verfasst von / submitted by Barbara Reischl, BSc angestrebter akademischer Grad / in partial fulfilment of the requirements for the degree of Master of Science (MSc) Wien, 2016 / Vienna 2016 Studienkennzahl lt. Studienblatt / A 066 830 degree programme code as it appears on the student record sheet: Studienrichtung lt. Studienblatt / Molecular Microbiology, Microbial Ecology degree programme as it appears on and Immunobiology the student record sheet: Betreut von / Supervisor: Univ.-Prof. Dipl.-Biol. Dr. Christa Schleper Mitbetreut von / Co-Supervisor: Mag. Mag. Dr. Simon Karl-Maria Rasso Rittmann, Bakk. 2 Table of Contents 1. Introduction .......................................................................................................... 5 1.1 Role of Biohydrogen ................................................................................................ 5 1.2 Hydrogenases ......................................................................................................... 6 1.3 Characteristics of Desulfurococcus fermentans ....................................................... 7 1.4 Aims of this Study ................................................................................................... 8 2. Material and Methods ............................................................................................ 9 2.1 Chemicals .............................................................................................................. -

Production of Bulk Chemicals with Biocatalysis: Drivers and Challenges Reflected in Recent Industrial Granted Patents
molecules Review Production of Bulk Chemicals with Biocatalysis: Drivers and Challenges Reflected in Recent Industrial Granted Patents (2015–2020) Nadia Guajardo 1,* and Pablo Domínguez de María 2 1 Programa Institucional de Fomento a la Investigación, Desarrollo e Innovación (PIDi), Universidad Tecnológica Metropolitana, Ignacio Valdivieso 2409, Santiago de Chile 8940000, Chile 2 Sustainable Momentum SL., Avenida Ansite 3, 4-6, 35011 Las Palmas de Gran Canaria, Spain; [email protected] * Correspondence: [email protected] Abstract: The application of biocatalysis and White Biotechnology tools in chemical areas concerning the production of bulk compounds and other related low-added value products (with high volumes) has been gaining importance in recent years. The expected drivers of biocatalysis for these sectors are energy savings, regioselectivity (leading to cleaner products), the possibility of using thermolabile substrates, as well as the generation of less by-products and manageable wastes. This paper explores some recent industrial granted patents related to biocatalysis and bulk chemicals. Several patents have been identified in fields such as biodiesel and esterification reactions, and sugar or furan chemistry. Overall, innovative strategies involve the identification of novel enzymes, the set-up of improved immobilization methods, as well as novel reactor designs that can offer improved performances and economics. The reported examples indicate that biocatalysis can certainly offer opportunities for Citation: Guajardo, N.; Domínguez these areas as well, far from the typical pharmaceutical and fine chemical applications often reported de María, P. Production of Bulk in the literature. Chemicals with Biocatalysis: Drivers and Challenges Reflected in Recent Keywords: biocatalysis; bulk compounds; sustainable processes; granted patents Industrial Granted Patents (2015–2020). -

Discovery of an O-Mannosylation Pathway Selectively Serving Cadherins and Protocadherins
Discovery of an O-mannosylation pathway selectively serving cadherins and protocadherins Ida Signe Bohse Larsena,1, Yoshiki Narimatsua,1, Hiren Jitendra Joshia,1, Lina Siukstaitea, Oliver J. Harrisonb, Julia Braschb, Kerry M. Goodmanb, Lars Hansena, Lawrence Shapirob,c,d, Barry Honigb,c,d,e, Sergey Y. Vakhrusheva, Henrik Clausena, and Adnan Halima,2,3 aDepartment of Cellular and Molecular Medicine, Faculty of Health Sciences, Copenhagen Center for Glycomics, University of Copenhagen, DK-2200 Copenhagen, Denmark; bDepartment of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032; cZuckerman Mind Brain Behavior Institute, Columbia University, New York, NY 10032; dDepartment of Systems Biology, Columbia University, New York, NY 10032; and eHoward Hughes Medical Institute, Columbia University, New York, NY 10032 Edited by Stuart A. Kornfeld, Washington University School of Medicine, St. Louis, MO, and approved September 6, 2017 (received for review May 22, 2017) The cadherin (cdh) superfamily of adhesion molecules carry muscular dystrophies that have been designated α-dystroglyca- O-linked mannose (O-Man) glycans at highly conserved sites nopathies because deficient O-Man glycosylation of α-DG dis- localized to specific β-strands of their extracellular cdh (EC) domains. rupts the interaction between the dystrophin glycoprotein complex These O-Man glycans do not appear to be elongated like O-Man and the ECM (7–9). Several studies have also implicated de- glycans found on α-dystroglycan (α-DG), and we recently demon- ficiency of POMT2 with E-cdh dysfunction (10–12), although di- strated that initiation of cdh/protocadherin (pcdh) O-Man glycosyl- rect evidence for a role in glycosylation of cdhs and pcdhs is ation is not dependent on the evolutionary conserved POMT1/ missing. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

The Rare Sugar D-Tagatose Protects Plants from Downy Mildews and Is a Safe Fungicidal Agrochemical
ARTICLE https://doi.org/10.1038/s42003-020-01133-7 OPEN The rare sugar D-tagatose protects plants from downy mildews and is a safe fungicidal agrochemical Susumu Mochizuki 1,3, Takeshi Fukumoto1,2,3, Toshiaki Ohara2,3, Kouhei Ohtani1, Akihide Yoshihara1, 1234567890():,; Yoshio Shigematsu 2, Keiji Tanaka2, Koichi Ebihara2, Shigeyuki Tajima1, Kenji Gomi1, Kazuya Ichimura 1, ✉ Ken Izumori1 & Kazuya Akimitsu 1 The rare sugar D-tagatose is a safe natural product used as a commercial food ingredient. Here, we show that D-tagatose controls a wide range of plant diseases and focus on downy mildews to analyze its mode of action. It likely acts directly on the pathogen, rather than as a plant defense activator. Synthesis of mannan and related products of D-mannose metabolism are essential for development of fungi and oomycetes; D-tagatose inhibits the first step of mannose metabolism, the phosphorylation of D-fructose to D-fructose 6-phosphate by fruc- tokinase, and also produces D-tagatose 6-phosphate. D-Tagatose 6-phosphate sequentially inhibits phosphomannose isomerase, causing a reduction in D-glucose 6-phosphate and D-fructose 6-phosphate, common substrates for glycolysis, and in D-mannose 6-phosphate, needed to synthesize mannan and related products. These chain-inhibitory effects on metabolic steps are significant enough to block initial infection and structural development needed for reproduction such as conidiophore and conidiospore formation of downy mildew. 1 International Institute of Rare Sugar Research and Education & Faculty of Agriculture, Kagawa University, 2393, Miki, Kagawa 761-0795, Japan. 2 Agrochemical Research Center, Mitsui Chemicals Agro, Inc., 1358 Ichimiyake, Yasu, Shiga 520-2362, Japan. -

(12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur Et Al
US 2012026.6329A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur et al. (43) Pub. Date: Oct. 18, 2012 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/10 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/24 (2006.01) CI2N 9/02 (2006.01) (75) Inventors: Eric J. Mathur, Carlsbad, CA CI2N 9/06 (2006.01) (US); Cathy Chang, San Marcos, CI2P 2L/02 (2006.01) CA (US) CI2O I/04 (2006.01) CI2N 9/96 (2006.01) (73) Assignee: BP Corporation North America CI2N 5/82 (2006.01) Inc., Houston, TX (US) CI2N 15/53 (2006.01) CI2N IS/54 (2006.01) CI2N 15/57 2006.O1 (22) Filed: Feb. 20, 2012 CI2N IS/60 308: Related U.S. Application Data EN f :08: (62) Division of application No. 1 1/817,403, filed on May AOIH 5/00 (2006.01) 7, 2008, now Pat. No. 8,119,385, filed as application AOIH 5/10 (2006.01) No. PCT/US2006/007642 on Mar. 3, 2006. C07K I4/00 (2006.01) CI2N IS/II (2006.01) (60) Provisional application No. 60/658,984, filed on Mar. AOIH I/06 (2006.01) 4, 2005. CI2N 15/63 (2006.01) Publication Classification (52) U.S. Cl. ................... 800/293; 435/320.1; 435/252.3: 435/325; 435/254.11: 435/254.2:435/348; (51) Int. Cl. 435/419; 435/195; 435/196; 435/198: 435/233; CI2N 15/52 (2006.01) 435/201:435/232; 435/208; 435/227; 435/193; CI2N 15/85 (2006.01) 435/200; 435/189: 435/191: 435/69.1; 435/34; CI2N 5/86 (2006.01) 435/188:536/23.2; 435/468; 800/298; 800/320; CI2N 15/867 (2006.01) 800/317.2: 800/317.4: 800/320.3: 800/306; CI2N 5/864 (2006.01) 800/312 800/320.2: 800/317.3; 800/322; CI2N 5/8 (2006.01) 800/320.1; 530/350, 536/23.1: 800/278; 800/294 CI2N I/2 (2006.01) CI2N 5/10 (2006.01) (57) ABSTRACT CI2N L/15 (2006.01) CI2N I/19 (2006.01) The invention provides polypeptides, including enzymes, CI2N 9/14 (2006.01) structural proteins and binding proteins, polynucleotides CI2N 9/16 (2006.01) encoding these polypeptides, and methods of making and CI2N 9/20 (2006.01) using these polynucleotides and polypeptides. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

The Impact of Endoplasmic Reticulum-Associated Protein Modifications, Folding and Degradation on Lung Structure and Function
REVIEW published: 25 May 2021 doi: 10.3389/fphys.2021.665622 The Impact of Endoplasmic Reticulum-Associated Protein Modifications, Folding and Degradation on Lung Structure and Function Emily M. Nakada1,2, Rui Sun1,2, Utako Fujii1,2 and James G. Martin1,2* 1Meakins-Christie Laboratories, Research Institute of the McGill University Health Centre (RI-MUHC), McGill University, Montreal, QC, Canada, 2McGill University, Montreal, QC, Canada The accumulation of unfolded/misfolded proteins in the endoplasmic reticulum (ER) causes ER stress and induces the unfolded protein response (UPR) and other mechanisms to restore ER homeostasis, including translational shutdown, increased targeting of mRNAs for degradation by the IRE1-dependent decay pathway, selective translation of proteins Edited by: that contribute to the protein folding capacity of the ER, and activation of the ER-associated Andrew John Halayko, degradation machinery. When ER stress is excessive or prolonged and these mechanisms University of Manitoba, Canada fail to restore proteostasis, the UPR triggers the cell to undergo apoptosis. This review Reviewed by: Amir A. Zeki, also examines the overlooked role of post-translational modifications and their roles in University of California, protein processing and effects on ER stress and the UPR. Finally, these effects are Davis, United States Pawan Sharma, examined in the context of lung structure, function, and disease. Thomas Jefferson University, Keywords: unfolded protein response, endoplasmic reticulum, integrated stress response, post-translational United States modifications, disulfide bonds, lung disease, lung function *Correspondence: James G. Martin [email protected] ENDOPLASMIC RETICULUM STRESS AND THE UNFOLDED Specialty section: PROTEIN RESPONSE This article was submitted to Respiratory Physiology, Cells are normally in a state of proteostasis, whereby networks of signaling pathways work in a section of the journal concert to maintain the proper synthesis, folding, trafficking, and degradation of proteins. -

(12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur Et Al
US 20150240226A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur et al. (43) Pub. Date: Aug. 27, 2015 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/16 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/02 (2006.01) CI2N 9/78 (2006.01) (71) Applicant: BP Corporation North America Inc., CI2N 9/12 (2006.01) Naperville, IL (US) CI2N 9/24 (2006.01) CI2O 1/02 (2006.01) (72) Inventors: Eric J. Mathur, San Diego, CA (US); CI2N 9/42 (2006.01) Cathy Chang, San Marcos, CA (US) (52) U.S. Cl. CPC. CI2N 9/88 (2013.01); C12O 1/02 (2013.01); (21) Appl. No.: 14/630,006 CI2O I/04 (2013.01): CI2N 9/80 (2013.01); CI2N 9/241.1 (2013.01); C12N 9/0065 (22) Filed: Feb. 24, 2015 (2013.01); C12N 9/2437 (2013.01); C12N 9/14 Related U.S. Application Data (2013.01); C12N 9/16 (2013.01); C12N 9/0061 (2013.01); C12N 9/78 (2013.01); C12N 9/0071 (62) Division of application No. 13/400,365, filed on Feb. (2013.01); C12N 9/1241 (2013.01): CI2N 20, 2012, now Pat. No. 8,962,800, which is a division 9/2482 (2013.01); C07K 2/00 (2013.01); C12Y of application No. 1 1/817,403, filed on May 7, 2008, 305/01004 (2013.01); C12Y 1 1 1/01016 now Pat. No. 8,119,385, filed as application No. PCT/ (2013.01); C12Y302/01004 (2013.01); C12Y US2006/007642 on Mar. 3, 2006. -

Supplementary Material (ESI) for Natural Product Reports
Electronic Supplementary Material (ESI) for Natural Product Reports. This journal is © The Royal Society of Chemistry 2014 Supplement to the paper of Alexey A. Lagunin, Rajesh K. Goel, Dinesh Y. Gawande, Priynka Pahwa, Tatyana A. Gloriozova, Alexander V. Dmitriev, Sergey M. Ivanov, Anastassia V. Rudik, Varvara I. Konova, Pavel V. Pogodin, Dmitry S. Druzhilovsky and Vladimir V. Poroikov “Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review” Contents PASS (Prediction of Activity Spectra for Substances) Approach S-1 Table S1. The lists of 122 known therapeutic effects for 50 analyzed medicinal plants with accuracy of PASS prediction calculated by a leave-one-out cross-validation procedure during the training and number of active compounds in PASS training set S-6 Table S2. The lists of 3,345 mechanisms of action that were predicted by PASS and were used in this study with accuracy of PASS prediction calculated by a leave-one-out cross-validation procedure during the training and number of active compounds in PASS training set S-9 Table S3. Comparison of direct PASS prediction results of known effects for phytoconstituents of 50 TIM plants with prediction of known effects through “mechanism-effect” and “target-pathway- effect” relationships from PharmaExpert S-79 S-1 PASS (Prediction of Activity Spectra for Substances) Approach PASS provides simultaneous predictions of many types of biological activity (activity spectrum) based on the structure of drug-like compounds. The approach used in PASS is based on the suggestion that biological activity of any drug-like compound is a function of its structure. -
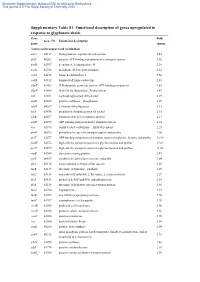
Supplementary Information Table S1. Up-Regulated Known Orfs Induced
Electronic Supplementary Material (ESI) for Molecular BioSystems This journal is © The Royal Society of Chemistry 2012 Supplementary Table S1. Functional description of genes upregulated in response to glyphosate shock Gene Fold Gene ID Functional description name change Amino acid transport and metabolism adiA b4117 biodegradative arginine decarboxylase 2.45 afuC b0262 putative ATP-binding component of a transport system 2.02 ansB b2957 periplasmic L-asparaginase II 2.28 argK b2918 membrane ATPase/protein kinase 2.12 cadA b4131 lysine decarboxylase 1 2.60 cadB b4132 transport of lysine cadaverine 2.83 ddpF b1483 D,D-dipeptide permease system, ATP-binding component 2.83 ddpX b1488 D-ala-D-ala dipeptidase, Zn-dependent 2.49 edd b1851 6-phosphogluconate dehydratase 2.29 elaD b2269 putative sulfatase phosphatase 2.07 idnD b4267 L-idonate dehydrogenase 2.31 nikA b3476 periplasmic binding protein for nickel 2.15 nikB b3477 transport of nickel, membrane protein 2.17 nikD b3479 ATP-binding protein of nickel transport system 2.24 oxc b2373 oxalyl CoA decarboxylase, ThDP-dependent 2.25 pheP b0576 phenylalanine-specific transport system and proline 2.08 proV b2677 ATP-binding component of transport system for glycine, betaine and proline 12.96 proW b2678 high-affinity transport system for glycine betaine and proline 17.69 proX b2679 high-affinity transport system for glycine betaine and proline 11.66 rspB b1580 starvation sensing protein 2.47 speF b0693 ornithine decarboxylase isozyme, inducible 3.04 tdcA b3118 transcriptional activator of tdc operon