Mouse Degs2 Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Data
Supplementary Fig. 1 A B Responder_Xenograft_ Responder_Xenograft_ NON- NON- Lu7336, Vehicle vs Lu7466, Vehicle vs Responder_Xenograft_ Responder_Xenograft_ Sagopilone, Welch- Sagopilone, Welch- Lu7187, Vehicle vs Lu7406, Vehicle vs Test: 638 Test: 600 Sagopilone, Welch- Sagopilone, Welch- Test: 468 Test: 482 Responder_Xenograft_ NON- Lu7860, Vehicle vs Responder_Xenograft_ Sagopilone, Welch - Lu7558, Vehicle vs Test: 605 Sagopilone, Welch- Test: 333 Supplementary Fig. 2 Supplementary Fig. 3 Supplementary Figure S1. Venn diagrams comparing probe sets regulated by Sagopilone treatment (10mg/kg for 24h) between individual models (Welsh Test ellipse p-value<0.001 or 5-fold change). A Sagopilone responder models, B Sagopilone non-responder models. Supplementary Figure S2. Pathway analysis of genes regulated by Sagopilone treatment in responder xenograft models 24h after Sagopilone treatment by GeneGo Metacore; the most significant pathway map representing cell cycle/spindle assembly and chromosome separation is shown, genes upregulated by Sagopilone treatment are marked with red thermometers. Supplementary Figure S3. GeneGo Metacore pathway analysis of genes differentially expressed between Sagopilone Responder and Non-Responder models displaying –log(p-Values) of most significant pathway maps. Supplementary Tables Supplementary Table 1. Response and activity in 22 non-small-cell lung cancer (NSCLC) xenograft models after treatment with Sagopilone and other cytotoxic agents commonly used in the management of NSCLC Tumor Model Response type -

DEGS2 Polymorphism Associated with Cognition in Schizophrenia Is Associated with Gene Expression in Brain
OPEN Citation: Transl Psychiatry (2015) 5, e550; doi:10.1038/tp.2015.45 www.nature.com/tp ORIGINAL ARTICLE DEGS2 polymorphism associated with cognition in schizophrenia is associated with gene expression in brain K Ohi1,2, G Ursini1,MLi1, JH Shin1,TYe1, Q Chen1,RTao1, JE Kleinman1, TM Hyde1,3,4, R Hashimoto2,5 and DR Weinberger1,3,4,6,7 A genome-wide association study of cognitive deficits in patients with schizophrenia in Japan found association with a missense genetic variant (rs7157599, Asn8Ser) in the delta(4)-desaturase, sphingolipid 2 (DEGS2) gene. A replication analysis using Caucasian samples showed a directionally consistent trend for cognitive association of a proxy single-nucleotide polymorphism (SNP), rs3783332. Although the DEGS2 gene is expressed in human brain, it is unknown how DEGS2 expression varies during human life and whether it is affected by psychiatric disorders and genetic variants. To address these questions, we examined DEGS2 messenger RNA using next-generation sequencing in postmortem dorsolateral prefrontal cortical tissue from a total of 418 Caucasian samples including patients with schizophrenia, bipolar disorder and major depressive disorder. DEGS2 is expressed at very low levels prenatally and increases gradually from birth to adolescence and consistently expressed across adulthood. Rs3783332 genotype −3 was significantly associated with the expression across all subjects (F3,348 = 10.79,P= 1.12 × 10 ), particularly in control subjects − 4 (F1,87 = 13.14, P = 4.86 × 10 ). Similar results were found with rs715799 genotype. The carriers of the risk-associated minor allele at both loci showed significantly lower expression compared with subjects homozygous for the non-risk major allele and this was a consistent finding across all diagnostic groups. -

Investigation of Adiposity Phenotypes in AA Associated with GALNT10 & Related Pathway Genes
Investigation of Adiposity Phenotypes in AA Associated With GALNT10 & Related Pathway Genes By Mary E. Stromberg A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY In Molecular Genetics and Genomics December 2018 Winston-Salem, North Carolina Approved by: Donald W. Bowden, Ph.D., Advisor Maggie C.Y. Ng, Ph.D., Advisor Timothy D. Howard, Ph.D., Chair Swapan Das, Ph.D. John P. Parks, Ph.D. Acknowledgements I would first like to thank my mentors, Dr. Bowden and Dr. Ng, for guiding my learning and growth during my years at Wake Forest University School of Medicine. Thank you Dr. Ng for spending so much time ensuring that I learn every detail of every protocol, and supporting me through personal difficulties over the years. Thank you Dr. Bowden for your guidance in making me a better scientist and person. I would like to thank my committee for their patience and the countless meetings we have had in discussing this project. I would like to say thank you to the members of our lab as well as the Parks lab for their support and friendship as well as their contributions to my project. Special thanks to Dean Godwin for his support and understanding. The umbrella program here at WFU has given me the chance to meet some of the best friends I could have wished for. I would like to also thank those who have taught me along the way and helped me to get to this point of my life, with special thanks to the late Dr. -
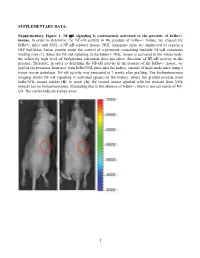
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

The Pdx1 Bound Swi/Snf Chromatin Remodeling Complex Regulates Pancreatic Progenitor Cell Proliferation and Mature Islet Β Cell
Page 1 of 125 Diabetes The Pdx1 bound Swi/Snf chromatin remodeling complex regulates pancreatic progenitor cell proliferation and mature islet β cell function Jason M. Spaeth1,2, Jin-Hua Liu1, Daniel Peters3, Min Guo1, Anna B. Osipovich1, Fardin Mohammadi3, Nilotpal Roy4, Anil Bhushan4, Mark A. Magnuson1, Matthias Hebrok4, Christopher V. E. Wright3, Roland Stein1,5 1 Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 2 Present address: Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN 3 Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN 4 Diabetes Center, Department of Medicine, UCSF, San Francisco, California 5 Corresponding author: [email protected]; (615)322-7026 1 Diabetes Publish Ahead of Print, published online June 14, 2019 Diabetes Page 2 of 125 Abstract Transcription factors positively and/or negatively impact gene expression by recruiting coregulatory factors, which interact through protein-protein binding. Here we demonstrate that mouse pancreas size and islet β cell function are controlled by the ATP-dependent Swi/Snf chromatin remodeling coregulatory complex that physically associates with Pdx1, a diabetes- linked transcription factor essential to pancreatic morphogenesis and adult islet-cell function and maintenance. Early embryonic deletion of just the Swi/Snf Brg1 ATPase subunit reduced multipotent pancreatic progenitor cell proliferation and resulted in pancreas hypoplasia. In contrast, removal of both Swi/Snf ATPase subunits, Brg1 and Brm, was necessary to compromise adult islet β cell activity, which included whole animal glucose intolerance, hyperglycemia and impaired insulin secretion. Notably, lineage-tracing analysis revealed Swi/Snf-deficient β cells lost the ability to produce the mRNAs for insulin and other key metabolic genes without effecting the expression of many essential islet-enriched transcription factors. -

Full Text (PDF)
medRxiv preprint doi: https://doi.org/10.1101/2020.12.29.20248986; this version posted January 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Insights into the molecular mechanism of anticancer drug ruxolitinib repurposable in COVID-19 therapy Manisha Mandal Department of Physiology, MGM Medical College, Kishanganj-855107, India Email: [email protected], ORCID: https://orcid.org/0000-0002-9562-5534 Shyamapada Mandal* Department of Zoology, University of Gour Banga, Malda-732103, India Email: [email protected], ORCID: https://orcid.org/0000-0002-9488-3523 *Corresponding author: Email: [email protected]; [email protected] Abstract Due to non-availability of specific therapeutics against COVID-19, repurposing of approved drugs is a reasonable option. Cytokines imbalance in COVID-19 resembles cancer; exploration of anti-inflammatory agents, might reduce COVID-19 mortality. The current study investigates the effect of ruxolitinib treatment in SARS-CoV-2 infected alveolar cells compared to the uninfected one from the GSE5147507 dataset. The protein-protein interaction network, biological process and functional enrichment of differentially expressed genes were studied using STRING App of the Cytoscape software and R programming tools. The present study indicated that ruxolitinib treatment elicited similar response equivalent to that of SARS-CoV-2 uninfected situation by inducing defense response in host against virus infection by RLR and NOD like receptor pathways. Further, the effect of ruxolitinib in SARS- CoV-2 infection was mainly caused by significant suppression of IFIH1, IRF7 and MX1 genes as well as inhibition of DDX58/IFIH1-mediated induction of interferon- I and -II signalling. -

Identification of MLL-AF9 Related Target Genes and Micrornas Involved in Leukemogenesis
Identification of MLL-AF9 related target genes and microRNAs involved in leukemogenesis Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) an der Fakultät für Biologie der Ludwig-Maximilians-Universität München Durchgeführt am Forschungszentrum des Dr. von Haunerschen Kinderspitals der Ludwig-Maximilians-Universität München vorgelegt von Katrin Kristina Fleischmann München, 3. Mai 2012 Erstgutachterin: Frau Prof. Dr. Elisabeth Weiss Zweitgutachter: Herr Prof. Dr. Michael Schleicher Sondergutachter: Herr Prof. Dr. Adelbert Roscher Tag der Abgabe: 03.05.2012 Tag der mündlichen Prüfung: 17.09.2012 What is a scientist after all? It is a curious man looking through a keyhole, the keyhole of nature, trying to know what's going on. Jacques Yves Cousteau Preamble and Acknowledgments How to explain the change from evolutionary ecology to tumor biology? After my diploma thesis in evolutionary ecology and a major in ecology, I was facing some skepticism while being interviewed for PhD positions in the biomedical field. What was my motivation to study biology? Why do I continue to feel this strong enthusiasm for this field? For me, it was always the grand question “How does life work?” that drove me and which may interconnect almost every aspect within natural sciences. Only when we strive to get answers to this grand question we may reach an understanding as to what happens in disease. I am profoundly grateful to my dear colleagues Dr. Julia von Frowein, Dr. Thomas Magg and Dr. Uta Fuchs who supported me to become one of their colleagues at the research center of the Dr. von Haunerschen Kinderspital. Since then, innumerable great and inspiring discussions allied us. -

Milger Et Al. Pulmonary CCR2+CD4+ T Cells Are Immune Regulatory And
Milger et al. Pulmonary CCR2+CD4+ T cells are immune regulatory and attenuate lung fibrosis development Supplemental Table S1 List of significantly regulated mRNAs between CCR2+ and CCR2- CD4+ Tcells on Affymetrix Mouse Gene ST 1.0 array. Genewise testing for differential expression by limma t-test and Benjamini-Hochberg multiple testing correction (FDR < 10%). Ratio, significant FDR<10% Probeset Gene symbol or ID Gene Title Entrez rawp BH (1680) 10590631 Ccr2 chemokine (C-C motif) receptor 2 12772 3.27E-09 1.33E-05 9.72 10547590 Klrg1 killer cell lectin-like receptor subfamily G, member 1 50928 1.17E-07 1.23E-04 6.57 10450154 H2-Aa histocompatibility 2, class II antigen A, alpha 14960 2.83E-07 1.71E-04 6.31 10590628 Ccr3 chemokine (C-C motif) receptor 3 12771 1.46E-07 1.30E-04 5.93 10519983 Fgl2 fibrinogen-like protein 2 14190 9.18E-08 1.09E-04 5.49 10349603 Il10 interleukin 10 16153 7.67E-06 1.29E-03 5.28 10590635 Ccr5 chemokine (C-C motif) receptor 5 /// chemokine (C-C motif) receptor 2 12774 5.64E-08 7.64E-05 5.02 10598013 Ccr5 chemokine (C-C motif) receptor 5 /// chemokine (C-C motif) receptor 2 12774 5.64E-08 7.64E-05 5.02 10475517 AA467197 expressed sequence AA467197 /// microRNA 147 433470 7.32E-04 2.68E-02 4.96 10503098 Lyn Yamaguchi sarcoma viral (v-yes-1) oncogene homolog 17096 3.98E-08 6.65E-05 4.89 10345791 Il1rl1 interleukin 1 receptor-like 1 17082 6.25E-08 8.08E-05 4.78 10580077 Rln3 relaxin 3 212108 7.77E-04 2.81E-02 4.77 10523156 Cxcl2 chemokine (C-X-C motif) ligand 2 20310 6.00E-04 2.35E-02 4.55 10456005 Cd74 CD74 antigen -

Downloaded from the Mouse Lysosome Gene Database, Mlgdb
1 Supplemental Figure Legends 2 3 Supplemental Figure S1: Epidermal-specific mTORC1 gain-of-function models show 4 increased mTORC1 activation and down-regulate EGFR and HER2 protein expression in a 5 mTORC1-sensitive manner. (A) Immunoblotting of Rheb1 S16H flox/flox keratinocyte cultures 6 infected with empty or adenoviral cre recombinase for markers of mTORC1 (p-S6, p-4E-BP1) 7 activity. (B) Tsc1 cKO epidermal lysates also show decreased expression of TSC2 by 8 immunoblotting of the same experiment as in Figure 2A. (C) Immunoblotting of Tsc2 flox/flox 9 keratinocyte cultures infected with empty or adenoviral cre recombinase showing decreased EGFR 10 and HER2 protein expression. (D) Expression of EGFR and HER2 was decreased in Tsc1 cre 11 keratinocytes compared to empty controls, and up-regulated in response to Torin1 (1µM, 24 hrs), 12 by immunoblot analyses. Immunoblots are contemporaneous and parallel from the same biological 13 replicate and represent the same experiment as depicted in Figure 7B. (E) Densitometry 14 quantification of representative immunoblot experiments shown in Figures 2E and S1D (r≥3; error 15 bars represent STDEV; p-values by Student’s T-test). 16 17 18 19 20 21 22 23 Supplemental Figure S2: EGFR and HER2 transcription are unchanged with epidermal/ 24 keratinocyte Tsc1 or Rptor loss. Egfr and Her2 mRNA levels in (A) Tsc1 cKO epidermal lysates, 25 (B) Tsc1 cKO keratinocyte lysates and(C) Tsc1 cre keratinocyte lysates are minimally altered 26 compared to their respective controls. (r≥3; error bars represent STDEV; p-values by Student’s T- 27 test). -

Kent Academic Repository Full Text Document (Pdf)
Kent Academic Repository Full text document (pdf) Citation for published version Alexander, Stephen PH and Fabbro, Doriano and Kelly, Eamonn and Mathie, Alistair and Peters, John A and Veale, Emma L. and Armstrong, Jane F and Faccenda, Elena and Harding, Simon D and Pawson, Adam J and Sharman, Joanna L and Southan, Christopher and Davies, Jamie A and Beuve, Annie and Boison, Detlev and Brouckaert, Peter and Burnett, John C and Burns, Kathryn DOI https://doi.org/10.1111/bph.14752 Link to record in KAR https://kar.kent.ac.uk/78599/ Document Version Publisher pdf Copyright & reuse Content in the Kent Academic Repository is made available for research purposes. Unless otherwise stated all content is protected by copyright and in the absence of an open licence (eg Creative Commons), permissions for further reuse of content should be sought from the publisher, author or other copyright holder. Versions of research The version in the Kent Academic Repository may differ from the final published version. Users are advised to check http://kar.kent.ac.uk for the status of the paper. Users should always cite the published version of record. Enquiries For any further enquiries regarding the licence status of this document, please contact: [email protected] If you believe this document infringes copyright then please contact the KAR admin team with the take-down information provided at http://kar.kent.ac.uk/contact.html S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology (2019) 176, S297–S396 -

Downloaded the HCV Specific Down-Regulated Genes Did Not Show Sig- from TCGA
Sun et al. BMC Medical Genomics (2019) 12:147 https://doi.org/10.1186/s12920-019-0580-x RESEARCH ARTICLE Open Access A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma Suofeng Sun1, Yuan Li2, Shuangyin Han1, Hongtao Jia3, Xiuling Li1* and Xiaofang Li1* Abstract Background: Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide, especially in East Asia. Even with the progress in therapy, 5-year survival rates remain unsatisfied. Chronic infection with the hepatitis B virus (HBV) or hepatitis C virus (HCV) has been epidemiologically associated with HCC and is the major etiology in the East Asian population. The detailed mechanism, especially the changes of DNA methylation and gene expression between the two types of virus-related HCC, and their contributions to the HCC development, metastasis, and recurrence remain largely unknown. Methods: In this integrated analysis, we characterized genome-scale profiles of HBV and HCV infected HCC by comparing their gene expression pattern, methylation profiles, and copy number variations from the publicly accessible data of The Cancer Genome Atlas Program (TCGA). Results: The HLA-A, STAT1, and OAS2 genes were highly enriched and up-regulated discovered in the HCV-infected HCC. Hypomethylation but not copy number variations might be the major factor for the up-regulation of these immune- related genes in HCV-infected HCC. Conclusions: The results indicated the different epigenetic changes of HBV/HCV related hepatocarcinogenesis. The top up-regulated genes in HCV group were significantly clustered in the immune-related and defense response pathways. These findings will help us to understand the pathogenesis of HBV/HCV associated hepatocellular carcinoma.