Use of Whole-Genome Sequencing to Diagnose a Cryptic Fusion Oncogene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Data
Supplementary Fig. 1 A B Responder_Xenograft_ Responder_Xenograft_ NON- NON- Lu7336, Vehicle vs Lu7466, Vehicle vs Responder_Xenograft_ Responder_Xenograft_ Sagopilone, Welch- Sagopilone, Welch- Lu7187, Vehicle vs Lu7406, Vehicle vs Test: 638 Test: 600 Sagopilone, Welch- Sagopilone, Welch- Test: 468 Test: 482 Responder_Xenograft_ NON- Lu7860, Vehicle vs Responder_Xenograft_ Sagopilone, Welch - Lu7558, Vehicle vs Test: 605 Sagopilone, Welch- Test: 333 Supplementary Fig. 2 Supplementary Fig. 3 Supplementary Figure S1. Venn diagrams comparing probe sets regulated by Sagopilone treatment (10mg/kg for 24h) between individual models (Welsh Test ellipse p-value<0.001 or 5-fold change). A Sagopilone responder models, B Sagopilone non-responder models. Supplementary Figure S2. Pathway analysis of genes regulated by Sagopilone treatment in responder xenograft models 24h after Sagopilone treatment by GeneGo Metacore; the most significant pathway map representing cell cycle/spindle assembly and chromosome separation is shown, genes upregulated by Sagopilone treatment are marked with red thermometers. Supplementary Figure S3. GeneGo Metacore pathway analysis of genes differentially expressed between Sagopilone Responder and Non-Responder models displaying –log(p-Values) of most significant pathway maps. Supplementary Tables Supplementary Table 1. Response and activity in 22 non-small-cell lung cancer (NSCLC) xenograft models after treatment with Sagopilone and other cytotoxic agents commonly used in the management of NSCLC Tumor Model Response type -

DEGS2 Polymorphism Associated with Cognition in Schizophrenia Is Associated with Gene Expression in Brain
OPEN Citation: Transl Psychiatry (2015) 5, e550; doi:10.1038/tp.2015.45 www.nature.com/tp ORIGINAL ARTICLE DEGS2 polymorphism associated with cognition in schizophrenia is associated with gene expression in brain K Ohi1,2, G Ursini1,MLi1, JH Shin1,TYe1, Q Chen1,RTao1, JE Kleinman1, TM Hyde1,3,4, R Hashimoto2,5 and DR Weinberger1,3,4,6,7 A genome-wide association study of cognitive deficits in patients with schizophrenia in Japan found association with a missense genetic variant (rs7157599, Asn8Ser) in the delta(4)-desaturase, sphingolipid 2 (DEGS2) gene. A replication analysis using Caucasian samples showed a directionally consistent trend for cognitive association of a proxy single-nucleotide polymorphism (SNP), rs3783332. Although the DEGS2 gene is expressed in human brain, it is unknown how DEGS2 expression varies during human life and whether it is affected by psychiatric disorders and genetic variants. To address these questions, we examined DEGS2 messenger RNA using next-generation sequencing in postmortem dorsolateral prefrontal cortical tissue from a total of 418 Caucasian samples including patients with schizophrenia, bipolar disorder and major depressive disorder. DEGS2 is expressed at very low levels prenatally and increases gradually from birth to adolescence and consistently expressed across adulthood. Rs3783332 genotype −3 was significantly associated with the expression across all subjects (F3,348 = 10.79,P= 1.12 × 10 ), particularly in control subjects − 4 (F1,87 = 13.14, P = 4.86 × 10 ). Similar results were found with rs715799 genotype. The carriers of the risk-associated minor allele at both loci showed significantly lower expression compared with subjects homozygous for the non-risk major allele and this was a consistent finding across all diagnostic groups. -

Investigation of Adiposity Phenotypes in AA Associated with GALNT10 & Related Pathway Genes
Investigation of Adiposity Phenotypes in AA Associated With GALNT10 & Related Pathway Genes By Mary E. Stromberg A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY In Molecular Genetics and Genomics December 2018 Winston-Salem, North Carolina Approved by: Donald W. Bowden, Ph.D., Advisor Maggie C.Y. Ng, Ph.D., Advisor Timothy D. Howard, Ph.D., Chair Swapan Das, Ph.D. John P. Parks, Ph.D. Acknowledgements I would first like to thank my mentors, Dr. Bowden and Dr. Ng, for guiding my learning and growth during my years at Wake Forest University School of Medicine. Thank you Dr. Ng for spending so much time ensuring that I learn every detail of every protocol, and supporting me through personal difficulties over the years. Thank you Dr. Bowden for your guidance in making me a better scientist and person. I would like to thank my committee for their patience and the countless meetings we have had in discussing this project. I would like to say thank you to the members of our lab as well as the Parks lab for their support and friendship as well as their contributions to my project. Special thanks to Dean Godwin for his support and understanding. The umbrella program here at WFU has given me the chance to meet some of the best friends I could have wished for. I would like to also thank those who have taught me along the way and helped me to get to this point of my life, with special thanks to the late Dr. -
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
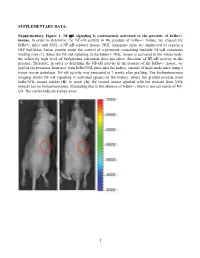
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

Ectopic Protein Interactions Within BRD4–Chromatin Complexes Drive Oncogenic Megadomain Formation in NUT Midline Carcinoma
Ectopic protein interactions within BRD4–chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma Artyom A. Alekseyenkoa,b,1, Erica M. Walshc,1, Barry M. Zeea,b, Tibor Pakozdid, Peter Hsic, Madeleine E. Lemieuxe, Paola Dal Cinc, Tan A. Incef,g,h,i, Peter V. Kharchenkod,j, Mitzi I. Kurodaa,b,2, and Christopher A. Frenchc,2 aDivision of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; bDepartment of Genetics, Harvard Medical School, Boston, MA 02115; cDepartment of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; dDepartment of Biomedical Informatics, Harvard Medical School, Boston, MA 02115; eBioinfo, Plantagenet, ON, Canada K0B 1L0; fDepartment of Pathology, University of Miami Miller School of Medicine, Miami, FL 33136; gBraman Family Breast Cancer Institute, University of Miami Miller School of Medicine, Miami, FL 33136; hInterdisciplinary Stem Cell Institute, University of Miami Miller School of Medicine, Miami, FL 33136; iSylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL 33136; and jHarvard Stem Cell Institute, Cambridge, MA 02138 Contributed by Mitzi I. Kuroda, April 6, 2017 (sent for review February 7, 2017; reviewed by Sharon Y. R. Dent and Jerry L. Workman) To investigate the mechanism that drives dramatic mistargeting of and, in the case of MYC, leads to differentiation in culture (2, 3). active chromatin in NUT midline carcinoma (NMC), we have Similarly, small-molecule BET inhibitors such as JQ1, which identified protein interactions unique to the BRD4–NUT fusion disengage BRD4–NUT from chromatin, diminish megadomain- oncoprotein compared with wild-type BRD4. -

Polyclonal Antibody to ZNF687
TA319985 OriGene Technologies Inc. OriGene EU Acris Antibodies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] Polyclonal Antibody to ZNF687 (C-term) - Aff - Purified Alternate names: KIAA1441, Zinc finger protein 687 Catalog No.: TA319985 Quantity: 0.1 mg Concentration: 1.0 mg/ml Background: The zinc finger protein 687 (ZNF687) was initially identified as a translocation partner gene with RUNX1 in patients with acute myeloid leukemia (AML). Little is known of the function of the ZNF687 protein, but it has been shown to weakly interact with the Ring1/Rnf2 RING finger protein member of the Polycomb group of proteins, suggesting it may be involved in the chromatin-modifying complexes essential for embryonic development and stem cell renewal. Other evidence suggests that ZNF687 may be part of a transcriptional network that also includes ZNF592 and ZMYMD8. Uniprot ID: Q8N1G0 NCBI: NP_065883 GeneID: 57592 Host / Isotype: Rabbit / IgG Immunogen: 17 amino acid synthetic peptide near the carboxy terminus of Human ZNF687 (AP55449CP- N) Format: State: Liquid purified Ig fraction Purification: Affinity chromatography purified via peptide column Buffer System: PBS containing 0.02% Sodium Azide as preservative Applications: Western blot: 0.5-1 µg/ml. Positive Control: Jurkat cell lysate. Immunofluorescence: Start at 20 μg/ml. Other applications not tested. Optimal dilutions are dependent on conditions and should be determined by the user. Specificity: This antibody is Human specific. -

ZNF687 CRISPR/Cas9 KO Plasmid (M): Sc-429638
SANTA CRUZ BIOTECHNOLOGY, INC. ZNF687 CRISPR/Cas9 KO Plasmid (m): sc-429638 BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and ZNF687 CRISPR/Cas9 KO Plasmid (m) is recommended for the disruption of CRISPR-associated protein (Cas9) system is an adaptive immune response gene expression in mouse cells. defense mechanism used by archea and bacteria for the degradation of foreign genetic material (4,6). This mechanism can be repurposed for other 20 nt non-coding RNA sequence: guides Cas9 functions, including genomic engineering for mammalian systems, such as to a specific target location in the genomic DNA gene knockout (KO) (1,2,3,5). CRISPR/Cas9 KO Plasmid products enable the U6 promoter: drives gRNA scaffold: helps Cas9 identification and cleavage of specific genes by utilizing guide RNA (gRNA) expression of gRNA bind to target DNA sequences derived from the Genome-scale CRISPR Knock-Out (GeCKO) v2 library developed in the Zhang Laboratory at the Broad Institute (3,5). Termination signal Green Fluorescent Protein: to visually REFERENCES verify transfection CRISPR/Cas9 Knockout Plasmid CBh (chicken β-Actin 1. Cong, L., et al. 2013. Multiplex genome engineering using CRISPR/Cas hybrid) promoter: drives systems. Science 339: 819-823. 2A peptide: expression of Cas9 allows production of both Cas9 and GFP from the 2. Mali, P., et al. 2013. RNA-guided human genome engineering via Cas9. same CBh promoter Science 339: 823-826. Nuclear localization signal 3. Ran, F.A., et al. 2013. Genome engineering using the CRISPR-Cas9 system. Nuclear localization signal SpCas9 ribonuclease Nat. Protoc. 8: 2281-2308. -

Atlas Journal
Atlas of Genetics and Cytogenetics in Oncology and Haematology Home Genes Leukemias Solid Tumours Cancer-Prone Deep Insight Portal Teaching X Y 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 NA Atlas Journal Atlas Journal versus Atlas Database: the accumulation of the issues of the Journal constitutes the body of the Database/Text-Book. TABLE OF CONTENTS Volume 12, Number 5, Sep-Oct 2008 Previous Issue / Next Issue Genes XAF1 (XIAP associated factor-1) (17p13.2). Stéphanie Plenchette, Wai Gin Fong, Robert G Korneluk. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 668-673. [Full Text] [PDF] URL : http://atlasgeneticsoncology.org/Genes/XAF1ID44095ch17p13.html WWP1 (WW domain containing E3 ubiquitin protein ligase 1) (8q21.3). Ceshi Chen. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 674-680. [Full Text] [PDF] URL : http://atlasgeneticsoncology.org/Genes/WWP1ID42993ch8q21.html TSPAN1 (tetraspanin 1) (1p34.1). David Murray, Peter Doran. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 681-683. [Full Text] [PDF] URL : http://atlasgeneticsoncology.org/Genes/TSPAN1ID44178ch1p34.html TCL1B (T-cell leukemia/lymphoma 1B) (14q32.13). Herbert Eradat, Michael A Teitell. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 684-686. [Full Text] [PDF] URL : http://atlasgeneticsoncology.org/Genes/TCL1BID354ch14q32.html PVRL4 (poliovirus receptor-related 4) (1q23.3). Marc Lopez. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 687-690. [Full Text] [PDF] URL : http://atlasgeneticsoncology.org/Genes/PVRL4ID44141ch1q23.html PTTG1IP (pituitary tumor-transforming 1 interacting protein) (21q22.3). Vicki Smith, Chris McCabe. Atlas Genet Cytogenet Oncol Haematol 2008; 12 (5): 691-694. -

Variation in Protein Coding Genes Identifies Information Flow
bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 1 1 2 3 4 5 Variation in protein coding genes identifies information flow as a contributor to 6 animal complexity 7 8 Jack Dean, Daniela Lopes Cardoso and Colin Sharpe* 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Institute of Biological and Biomedical Sciences 25 School of Biological Science 26 University of Portsmouth, 27 Portsmouth, UK 28 PO16 7YH 29 30 * Author for correspondence 31 [email protected] 32 33 Orcid numbers: 34 DLC: 0000-0003-2683-1745 35 CS: 0000-0002-5022-0840 36 37 38 39 40 41 42 43 44 45 46 47 48 49 Abstract bioRxiv preprint doi: https://doi.org/10.1101/679456; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Animal complexity and information flow 2 1 Across the metazoans there is a trend towards greater organismal complexity. How 2 complexity is generated, however, is uncertain. Since C.elegans and humans have 3 approximately the same number of genes, the explanation will depend on how genes are 4 used, rather than their absolute number. -

Chromatin Conformation Links Distal Target Genes to CKD Loci
BASIC RESEARCH www.jasn.org Chromatin Conformation Links Distal Target Genes to CKD Loci Maarten M. Brandt,1 Claartje A. Meddens,2,3 Laura Louzao-Martinez,4 Noortje A.M. van den Dungen,5,6 Nico R. Lansu,2,3,6 Edward E.S. Nieuwenhuis,2 Dirk J. Duncker,1 Marianne C. Verhaar,4 Jaap A. Joles,4 Michal Mokry,2,3,6 and Caroline Cheng1,4 1Experimental Cardiology, Department of Cardiology, Thoraxcenter Erasmus University Medical Center, Rotterdam, The Netherlands; and 2Department of Pediatrics, Wilhelmina Children’s Hospital, 3Regenerative Medicine Center Utrecht, Department of Pediatrics, 4Department of Nephrology and Hypertension, Division of Internal Medicine and Dermatology, 5Department of Cardiology, Division Heart and Lungs, and 6Epigenomics Facility, Department of Cardiology, University Medical Center Utrecht, Utrecht, The Netherlands ABSTRACT Genome-wide association studies (GWASs) have identified many genetic risk factors for CKD. However, linking common variants to genes that are causal for CKD etiology remains challenging. By adapting self-transcribing active regulatory region sequencing, we evaluated the effect of genetic variation on DNA regulatory elements (DREs). Variants in linkage with the CKD-associated single-nucleotide polymorphism rs11959928 were shown to affect DRE function, illustrating that genes regulated by DREs colocalizing with CKD-associated variation can be dysregulated and therefore, considered as CKD candidate genes. To identify target genes of these DREs, we used circular chro- mosome conformation capture (4C) sequencing on glomerular endothelial cells and renal tubular epithelial cells. Our 4C analyses revealed interactions of CKD-associated susceptibility regions with the transcriptional start sites of 304 target genes. Overlap with multiple databases confirmed that many of these target genes are involved in kidney homeostasis. -

1 Genome-Wide Discovery of SLE Genetic Risk Variant Allelic Enhancer
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.20.906701; this version posted January 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Genome-wide discovery of SLE genetic risk variant allelic enhancer activity Xiaoming Lu*1, Xiaoting Chen*1, Carmy Forney1, Omer Donmez1, Daniel Miller1, Sreeja Parameswaran1, Ted Hong1,2, Yongbo Huang1, Mario Pujato3, Tareian Cazares4, Emily R. Miraldi3-5, John P. Ray6, Carl G. de Boer6, John B. Harley1,4,5,7,8, Matthew T. Weirauch#,1,3,5,8,9, Leah C. Kottyan#,1,4,5,9 *Contributed equally #Co-corresponding authors: [email protected]; [email protected] 1Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA, 45229. 2Department of Pharmacology & Systems Physiology, University of Cincinnati, College of Medicine, Cincinnati, Ohio, USA, 45229. 3Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA, 45229. 4Division of Immunobiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA, 45229. 5Department of Pediatrics, University of Cincinnati, College of Medicine, Cincinnati, Ohio, USA, 45229. 6Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard University, Cambridge, Massachusetts, USA, 02142. 7US Department of Veterans Affairs Medical Center, Cincinnati, Ohio, USA 45229. 8Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA, 45229. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.01.20.906701; this version posted January 20, 2020. -

Agricultural University of Athens
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΩΝ ΖΩΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΖΩΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΝΙΚΗΣ ΚΑΙ ΕΙΔΙΚΗΣ ΖΩΟΤΕΧΝΙΑΣ ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ ΑΘΗΝΑ 2020 ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Genome-wide association analysis and gene network analysis for (re)production traits in commercial broilers ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ Τριμελής Επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Επταμελής εξεταστική επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Πηνελόπη Μπεμπέλη (Καθ. ΓΠΑ) Δημήτριος Βλαχάκης (Επ. Καθ. ΓΠΑ) Ευάγγελος Ζωίδης (Επ.Καθ. ΓΠΑ) Γεώργιος Θεοδώρου (Επ.Καθ. ΓΠΑ) 2 Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Περίληψη Σκοπός της παρούσας διδακτορικής διατριβής ήταν ο εντοπισμός γενετικών δεικτών και υποψηφίων γονιδίων που εμπλέκονται στο γενετικό έλεγχο δύο τυπικών πολυγονιδιακών ιδιοτήτων σε κρεοπαραγωγικά ορνίθια. Μία ιδιότητα σχετίζεται με την ανάπτυξη (σωματικό βάρος στις 35 ημέρες, ΣΒ) και η άλλη με την αναπαραγωγική