Pyridine-2-Carboximidamidate Chloride Monohydrate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2017 Annual Report 1 Definitions
* Bank of Jinzhou Co., Ltd. is not an authorized institution within the meaning of the Banking Ordinane (Chapter 155 of the Laws of Hong Kong), not subject to the supervision of the Hong Kong Monetary Authority, and not authorized to carry on banking and/or deposit-taking business in Hong Kong. Contents 2 Definitions 4 Chapter 1 Company Profile 7 Chapter 2 Financial Highlights 10 Chapter 3 Chairman ’s Statement 12 Chapter 4 President’s Statement 14 Chapter 5 Management Discussion and Analysis 71 Chapter 6 Changes in Ordinary Shares and Particulars of Shareholders 77 Chapter 7 Particulars of Preference Shares 79 Chapter 8 Directors, Supervisors, Senior Management, Employees and Organizations 98 Chapter 9 Corporate Governance Report 119 Chapter 10 Directors’ Report 127 Chapter 11 Supervisors’ Report 130 Chapter 12 Social Responsibility Report 132 Chapter 13 Internal Control and Internal Audit 136 Chapter 14 Important Events 139 Chapter 15 Independent Auditor’s Report 149 Chapter 16 Financial Statements 269 Chapter 17 Unaudited Supplementary Financial Information Bank of Jinzhou Co., Ltd. 2017 Annual Report 1 Definitions In this annual report, unless the context otherwise requires, the following terms shall have the meanings set out below: “A Share Offering” the Bank’s proposed initial public offering of not more than 1,927,000,000 A shares, which has been approved by the Shareholders on 29 June 2016 “Articles of Association” the articles of association of the Bank, as the same may be amended from time to time “the Bank”, “Bank of Jinzhou” -

Table of Codes for Each Court of Each Level
Table of Codes for Each Court of Each Level Corresponding Type Chinese Court Region Court Name Administrative Name Code Code Area Supreme People’s Court 最高人民法院 最高法 Higher People's Court of 北京市高级人民 Beijing 京 110000 1 Beijing Municipality 法院 Municipality No. 1 Intermediate People's 北京市第一中级 京 01 2 Court of Beijing Municipality 人民法院 Shijingshan Shijingshan District People’s 北京市石景山区 京 0107 110107 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Haidian District of Haidian District People’s 北京市海淀区人 京 0108 110108 Beijing 1 Court of Beijing Municipality 民法院 Municipality Mentougou Mentougou District People’s 北京市门头沟区 京 0109 110109 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Changping Changping District People’s 北京市昌平区人 京 0114 110114 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Yanqing County People’s 延庆县人民法院 京 0229 110229 Yanqing County 1 Court No. 2 Intermediate People's 北京市第二中级 京 02 2 Court of Beijing Municipality 人民法院 Dongcheng Dongcheng District People’s 北京市东城区人 京 0101 110101 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Xicheng District Xicheng District People’s 北京市西城区人 京 0102 110102 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Fengtai District of Fengtai District People’s 北京市丰台区人 京 0106 110106 Beijing 1 Court of Beijing Municipality 民法院 Municipality 1 Fangshan District Fangshan District People’s 北京市房山区人 京 0111 110111 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Daxing District of Daxing District People’s 北京市大兴区人 京 0115 -

Annual Report 2019 Annual Report
Annual Report 2019 Annual Report 2019 For more information, please refer to : CONTENTS DEFINITIONS 2 Section I Important Notes 5 Section II Company Profile and Major Financial Information 6 Section III Company Business Overview 18 Section IV Discussion and Analysis on Operation 22 Section V Directors’ Report 61 Section VI Other Significant Events 76 Section VII Changes in Shares and Information on Shareholders 93 Section VIII Directors, Supervisors, Senior Management and Staff 99 Section IX Corporate Governance Report 119 Section X Independent Auditor’s Report 145 Section XI Consolidated Financial Statements 151 Appendix I Information on Securities Branches 276 Appendix II Information on Branch Offices 306 China Galaxy Securities Co., Ltd. Annual Report 2019 1 DEFINITIONS “A Share(s)” domestic shares in the share capital of the Company with a nominal value of RMB1.00 each, which is (are) listed on the SSE, subscribed for and traded in Renminbi “Articles of Association” the articles of association of the Company (as amended from time to time) “Board” or “Board of Directors” the board of Directors of the Company “CG Code” Corporate Governance Code and Corporate Governance Report set out in Appendix 14 to the Stock Exchange Listing Rules “Company”, “we” or “us” China Galaxy Securities Co., Ltd.(中國銀河證券股份有限公司), a joint stock limited company incorporated in the PRC on 26 January 2007, whose H Shares are listed on the Hong Kong Stock Exchange (Stock Code: 06881), the A Shares of which are listed on the SSE (Stock Code: 601881) “Company Law” -

Minimum Wage Standards in China August 11, 2020
Minimum Wage Standards in China August 11, 2020 Contents Heilongjiang ................................................................................................................................................. 3 Jilin ............................................................................................................................................................... 3 Liaoning ........................................................................................................................................................ 4 Inner Mongolia Autonomous Region ........................................................................................................... 7 Beijing......................................................................................................................................................... 10 Hebei ........................................................................................................................................................... 11 Henan .......................................................................................................................................................... 13 Shandong .................................................................................................................................................... 14 Shanxi ......................................................................................................................................................... 16 Shaanxi ...................................................................................................................................................... -

Area B ANSHAN FUYU ARTWORK CO., LTD
第118届 第2期 the 118th session Phase 2 温馨提示 “*”表示参展出口企业有内销意愿 ( Note):“*”means the exhibitor has the intention of domestic sale 1.1A01 依次表示为1号馆1楼A通道01号摊位 B 1.1A01 Orderly Indicates Hall No. 1, Corridor No. A and the Booth number "01" 区 (10.1A15-16)广州市伟冠贸易有限公司 礼品及赠品 GUANGZHOU WEI GUAN TRADE CO., LTD. Gifts And Premiums 广州市荔湾区东漖北路436号力诚凯怡大厦503室 (邮政编码:510370) 礼品及赠品 ROOM 503, LICHENGKAIYI INTERNATIONAL BUILDING, NO.436, (10.1A01-03)中艺家居制品(北京)有限公司 DONGJIAO NORTH ROAD, LIWAN DISTRICT, GUANGZHOU, CHINA : : CHINA FURNITURE CO., LTD. TEL 86-20-81237390 FAX 86-20-81237394 URL:WWW.WEIGUANTRADE.COM 北京朝阳门外吉祥里103号中国工艺大厦704 E-MAIL:[email protected] (邮政编码:100020) RM.704, CHINA ARTS & CRAFTS BUILDING, NO.103 JIXIANG (10.1A17-18)泉州雯婷艺品有限公司 LANE, CHAOYANGMENWAI, BEIJING, CHINA QUANZHOU WENTING CRAFTS CO., LTD. TEL:86-10-85698594 FAX:86-10-85698598 泉州市丰泽区城东镇新前村1号 E-MAIL:[email protected] (邮政编码:362000) (10.1A04-05)鞍山福宇工艺品有限公司 NO.1 XINQIAN, CHENGDONG, FENGZE DIST., QUANZHOU, FUJIAN, CHINA B Area ANSHAN FUYU ARTWORK CO., LTD. TEL:86-595-22686339 FAX:86-595-22678339 铁东区南胜利路21号万科写字楼1701-02室 E-MAIL:[email protected] (邮政编码:114001) ROOM 1701-02, VANKE OFFICE BUILDING, NO.21 SHENGLI *(10.1A19-20)上海经茂实业有限责任公司 SOUTH ROAD, TIEDONG DISTRICT, ANSHAN, LIAONING, CHINA SHANGHAI JINGMAO INDUSTRIAL CO., LTD. TEL:86-412-5558510 FAX:86-412-5558560 上海市浦东新区海徐路939号231室 URL:WWW.FUYU-FINE.COM (邮政编码:200433) Gifts And Premiums E-MAIL:[email protected] RM.231, NO.939 HAIXU ROAD, PUDONG NEW DIST., SHANGHAI TEL:86-21-60956956 FAX:86-21-60956950 (10.1A06-10)泉州奎生工艺有限公司 URL:WWW.JM-CN.COM QUANZHOU KUISHENG CRAFT CO., LTD. E-MAIL:[email protected] 泉州市丰泽区浔美工业区奎生工业园 (邮政编码:362000) (10.1B01-02)汕头市嘉鸿工艺厂有限公司 KUISHENG INDUSTRIAL PARK, XUNMEI INDUSTRIAL ZONE, SHANTOU JIAHONG CRAFT FACTORY CO., LTD. -

Area Comprehensive Score 1990 2000 2010 Heping District 0.307
Comprehensive score of aging level in 1990, 2000 and 2010 Comprehensive score Area 1990 2000 2010 Heping District 0.307 0.572 0.792 Shenhe District 0.319 0.554 0.774 Dadong District 0.275 0.558 0.803 Huanggu District 0.262 0.542 0.777 Tiexi District (Shenyang) 0.252 0.611 0.800 Sujiatun District 0.202 0.409 0.699 Dongling District 0.202 0.370 0.512 Shenbei New District 0.196 0.388 0.534 Yuhong District 0.197 0.364 0.593 Liaozhong County 0.187 0.351 0.627 Kangping County 0.165 0.318 0.604 Faku County 0.195 0.354 0.653 Xinmin City 0.177 0.351 0.627 Zhongshan District 0.336 0.592 0.888 Xigang District 0.327 0.605 0.860 Shahekou District 0.284 0.534 0.770 Ganjingzi District 0.242 0.381 0.557 Lushunkou District 0.302 0.427 0.668 Jinzhou District 0.267 0.360 0.531 Changhai County 0.215 0.314 0.638 Wafangdian City 0.218 0.431 0.799 Pulandian City 0.243 0.440 0.812 Zhuanghe City 0.224 0.460 0.778 Tiedong District 0.230 0.541 0.831 Tiexi District (Anshan) 0.234 0.514 0.896 Lishan District 0.198 0.540 0.950 Qianshan District 0.215 0.399 0.721 Tai'an County 0.187 0.355 0.613 Xiuyan Manchu Autonomous County 0.171 0.349 0.620 Haicheng City 0.191 0.321 0.573 Xinfu District 0.245 0.517 0.853 Dongzhou District 0.230 0.551 1.000 Wanghua District 0.206 0.464 0.814 Shuncheng District 0.195 0.479 0.819 Fushun County 0.256 0.401 0.701 Xinbin Manchu Autonomous County 0.110 0.298 0.615 Qingyuan Manchu Autonomous County 0.124 0.318 0.618 Pingshan District 0.208 0.475 0.778 Xihu District 0.217 0.497 0.829 Mingshan District 0.186 0.440 0.743 Nanfen District 0.196 -
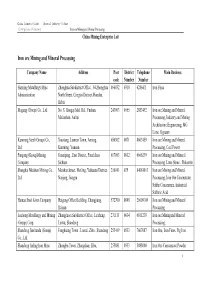
Iron Ore Mining and Mineral Processing China Mining Enterprise Lsit
China Commerce Guide -- Mineral Industry Volume Enterprises Directory Iron ore Mining and Mineral Processing China Mining Enterprise Lsit Iron ore Mining and Mineral Processing Company Name Address Post District Telephone Main Business code Number Number Hanxing Metallurgy Mine Zhonghua Subdistrict Office , 54 Zhonghua 056032 0310 6286411 Iron Fines Administration North Street, Congtai District, Handan, Hebei Magang (Group) Co., Ltd. No. 8, Hongqi Mid. Rd., Yushan, 243003 0555 2883492 Iron ore Mining and Mineral Ma'anshan, Anhui Processing, Industry and Mining Architecture Engineering, MG Lime, Gypsum Kunming Steel (Group) Co., Xiaotang, Lianran Town, Anning, 650302 0871 8603459 Iron ore Mining and Mineral Ltd. Kunming, Yunnan Processing, Coal Power Pangang (Group)Mining Guaziping , East District, Panzhihua, 617063 0812 6666239 Iron ore Mining and Mineral Company Sichuan Processing, Lime Stone,Dolomite Shanghai Meishan Mining Co., Meishan Street, Meiling, Yuhuatai District, 210041 025 84084013 Iron ore Mining and Mineral Ltd. Nanjing, Jiangsu Processing, Iron Ore Concentrate, Sulfur Concentrate, Industrial Sulfuric Acid Hainan Steel & iron Company Haigang Office Building, Changjiang, 572700 0898 26609109 Iron ore Mining and Mineral Hainan Processing Luzhong Metallurgy and Mining Zhangjiawa Subdistrict Office, Laicheng, 271113 0634 6811238 Iron ore Miningand Mineral (Group) Corp. Laiwu, Shandong Processing; Shandong Jinshunda (Group) Fenghuang Town , Lionzi, Zibo , Shandong 255419 0533 7607087 Iron Ore, Iron Fines, Pig Iron Co., Ltd. Shandong -
2018 China Adult Tobacco Survey Report
2018 China Adult Tobacco Survey Report Contents Definition of indicators ................................................................................................................................... 5 Chapter 1 Introduction .................................................................................................................................... 6 Chapter 2 Survey methodology ...................................................................................................................... 9 2.1 Goals of study ...................................................................................................................................... 9 2.2 Target population ................................................................................................................................. 9 2.3 Inclusion criteria .................................................................................................................................. 9 2.4 Sampling design .................................................................................................................................. 9 2.5 Questionnaire survey .........................................................................................................................11 2.6 Data collection ...................................................................................................................................11 2.7 Statistical analysis .............................................................................................................................12 -

Notice of OFAC Sanctions Actions AGENCY
This document is scheduled to be published in the Federal Register on 10/20/2020 and available online at federalregister.gov/d/2020-22980, and on govinfo.gov DEPARTMENT OF THE TREASURY Office of Foreign Assets Control [Case ID DPRK-17839] Notice of OFAC Sanctions Actions AGENCY: Office of Foreign Assets Control, Treasury. ACTION: Notice. SUMMARY: The Department of the Treasury’s Office of Foreign Assets Control (OFAC) is updating the entries of 490 persons on OFAC’s Specially Designated Nationals and Blocked Persons List (SDN List). DATES: See Supplementary Information section for applicable date(s). FOR FURTHER INFORMATION CONTACT: OFAC: Associate Director for Global Targeting, tel.: 202-622-2420; Assistant Director for Licensing, tel.: 202-622-2480; Assistant Director for Regulatory Affairs, tel.: 202-622-4855; Assistant Director for Sanctions Compliance & Evaluation, tel.: 202-622-2490; or the Department of the Treasury’s Office of the General Counsel: Office of the Chief Counsel (Foreign Assets Control), tel.: 202-622-2410. SUPPLEMENTARY INFORMATION: Electronic Availability The Specially Designated Nationals and Blocked Persons List and additional information concerning OFAC sanctions programs are available on OFAC’s website (www.treasury.gov/ofac). Notice of OFAC Actions On April 10, 2020 OFAC amended the North Korea Sanctions Regulations, 31 CFR part 510, to implement the Treasury-administered provisions of the North Korea Sanctions and Policy Enhancement Act of 2016, as amended by the Countering America’s Adversaries Through Sanctions Act and the National Defense Authorization Act for Fiscal Year 2020. Specifically, OFAC added a new prohibition to the regulations that is applicable to persons that are owned or controlled by a U.S. -

Minimum Wage Standards in China June 28, 2018
Minimum Wage Standards in China June 28, 2018 Contents Heilongjiang .................................................................................................................................................. 3 Jilin ................................................................................................................................................................ 3 Liaoning ........................................................................................................................................................ 4 Inner Mongolia Autonomous Region ........................................................................................................... 7 Beijing ......................................................................................................................................................... 10 Hebei ........................................................................................................................................................... 11 Henan .......................................................................................................................................................... 13 Shandong .................................................................................................................................................... 14 Shanxi ......................................................................................................................................................... 16 Shaanxi ....................................................................................................................................................... -

Annual Development Report on China's Trademark Strategy 2013
Annual Development Report on China's Trademark Strategy 2013 TRADEMARK OFFICE/TRADEMARK REVIEW AND ADJUDICATION BOARD OF STATE ADMINISTRATION FOR INDUSTRY AND COMMERCE PEOPLE’S REPUBLIC OF CHINA China Industry & Commerce Press Preface Preface 2013 was a crucial year for comprehensively implementing the conclusions of the 18th CPC National Congress and the second & third plenary session of the 18th CPC Central Committee. Facing the new situation and task of thoroughly reforming and duty transformation, as well as the opportunities and challenges brought by the revised Trademark Law, Trademark staff in AICs at all levels followed the arrangement of SAIC and got new achievements by carrying out trademark strategy and taking innovation on trademark practice, theory and mechanism. ——Trademark examination and review achieved great progress. In 2013, trademark applications increased to 1.8815 million, with a year-on-year growth of 14.15%, reaching a new record in the history and keeping the highest a mount of the world for consecutive 12 years. Under the pressure of trademark examination, Trademark Office and TRAB of SAIC faced the difficuties positively, and made great efforts on soloving problems. Trademark Office and TRAB of SAIC optimized the examination procedure, properly allocated examiners, implemented the mechanism of performance incentive, and carried out the “double-points” management. As a result, the Office examined 1.4246 million trademark applications, 16.09% more than last year. The examination period was maintained within 10 months, and opposition period was shortened to 12 months, which laid a firm foundation for performing the statutory time limit. —— Implementing trademark strategy with a shift to effective use and protection of trademark by law. -

The Role of Japan in Modern Chinese Art Edited by Joshua A
The Role of Japan in Modern Chinese Art Edited by Joshua A. Fogel Published in association with the University of California Press “This ambitious, very important project defines no less than a new field of inquiry, one that scarcely could have been attempted in the past. The essays in this volume add enor- mously to the documentation of what late-period Chinese art learned from Japan, and begin to formulate conclu- sions that will enrich future accounts of both Japanese and Chinese art.” JAMES CAHILL, University of California, Berkeley The modern histories of China and Japan are inexorably intertwined. Their relationship is perhaps most obvious in the fields of political, economic, and military history, but it is no less true in cultural and art history. Yet the traffic in artistic practices and practitioners between China and Japan remains an understudied field. In this volume, an international group of scholars investigates Japan’s impact on Chinese art from the mid-nineteenth cen- tury through the 1930s. Individual essays address a range of perspectives, including the work of individual Chinese and Japanese painters, calligraphers, and sculptors, as well as artistic associations, international exhibitions, the collotype production or artwork, and the emergence of a modern canon. JOSHUA A. FOGEL is Canada Research Chair and a professor of history at York University, Toron - to, and a specialist in the history of cultural and political ties between China and Japan in the mod - ern era. CONTRIBUTORS: Julia F. Andrews | Shana J. Brown | Chen Jie | Lisa Claypool | Walter B. Davis | Zaixin Hong | Yu-chih Lai | Tamaki Maeda | Kuiyi Shen | Richard Vinograd | Cheng-hua Wang | Aida Yuen Wong New Perspectives on Chinese Culture and Society, 3 The Role of Japan in Modern Chinese Art New PersPectives oN chiNese culture aNd society A series sponsored by the American Council of Learned Societies and made possible through a grant from the Chiang Ching-kuo Foundation for International Scholarly Exchange 1.