Supplementary Table 1. List of Proteins Quantitated in Tumor Tissue Treated with Versus Without IPI-504 Itraq Ratio Itraq Ratio Refseq (Rx/Norx) (Rx/Norx)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ZMYND10 Functions in a Chaperone Relay During Axonemal Dynein Assembly
bioRxiv preprint doi: https://doi.org/10.1101/233718; this version posted December 13, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 ZMYND10 functions in a chaperone relay during axonemal dynein assembly. 2 3 Girish R Mali1,9 , Patricia Yeyati1, Seiya Mizuno2, Margaret A Keighren1, Petra zur Lage3, Amaya 4 Garcia-Munoz4, Atsuko Shimada5, Hiroyuki Takeda5, Frank Edlich6, Satoru Takahashi2,7, Alex von 5 Kreigsheim4,8, Andrew Jarman3 and Pleasantine Mill1,*. 6 7 1. MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of 8 Edinburgh, Edinburgh, UK, EH4 2XU 9 2. Laboratory Animal Resource Centre, University of Tsukuba, Tsukuba, Japan, 305-8575 10 3. Centre for Integrative Physiology, University of Edinburgh, Edinburgh, UK, EH8 9XD 11 4. Systems Biology Ireland, University College Dublin, Dublin, Ireland 12 5. Department of Biological Sciences, University of Tokyo, Tokyo, Japan, 113-0033 13 6. Institute for Biochemistry and Molecular Biology, University of Freiburg, Freiburg, Germany, 14 79104 15 7. Department of Anatomy and Embryology, Faculty of Medicine, University of Tsukuba, Tsukuba, 16 Japan, 305-8575 17 8. Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University 18 of Edinburgh, Edinburgh, UK, EH4 2XU 19 9. Current address: MRC Laboratory of Molecular Biology, Cambridge, UK, CB2 0QH 20 * Corresponding author: [email protected] 21 22 23 24 25 26 27 28 29 30 31 1 bioRxiv preprint doi: https://doi.org/10.1101/233718; this version posted December 13, 2017. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

Supplementary Materials Functional Characterization of Rare RAB12
1 Supplementary materials Functional characterization of rare RAB12 variants and their role in musician’s and other dystonias Eva Hebert et al. Figure S1. Photograph of Individual L-10289 (mildly affected mother of the index patient from Family D) showing a 15-degree tilt of the trunk to the right as well as dystonic posturing of the right hand (involuntary flexion of the third to fifth finger and thumb and extension of the index finger). 2 Figure S2. TFRC colocalized with wildtype and mutant FLAG-RAB12. Immunofluorescent staining of fibroblasts expressing FLAG-RAB12 WT, p.Gly13Asp, or p.Ile196Val revealed predominant perinuclear localization of TFRC (red) which overlaps with the localization of FLAG-RAB12 (green) in all three cell lines (WT, p.Gly13Asp, p.Ile196Val). The nucleus was stained with DAPI (blue). Scale bar: 20µm. 3 Figure S3. Lysosomal degradation of the physiological dimeric TFRC was not affected by the RAB12 mutations. Western Blot analysis revealed the degradation of TFRC in patient fibroblasts with endogenous expression of RAB12 (a, b) in fibroblasts ectopically expressing FLAG-RAB12 (c, d), and in SH-SY5Y cells ectopically expressing FLAG-RAB12 (e, f). Cells were treated with Bafilomycin A1 for 24h. ß-actin served as loading control and for normalization. Bars in B, D, and F indicate means of three independent experiments ± SEM. ctrl control, WT wildtype 4 Figure S4. Relative LC3-II protein levels are marginally increased in SH-SY5Y cells overexpressing RAB12 Gly13Asp protein and p62 levels remained constant. a) Western Blot of proteins extracted from stably transfected SH-SY5Y cells. Expression of FLAG-tagged RAB12 WT equals the expression of mutated RAB12 proteins (Gly13Asp, I196Val) (lane 3, 5, 7). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

1 Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental
Page 1 of 255 Diabetes Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy Oliver J. Freeman1,2, Richard D. Unwin2,3, Andrew W. Dowsey2,3, Paul Begley2,3, Sumia Ali1, Katherine A. Hollywood2,3, Nitin Rustogi2,3, Rasmus S. Petersen1, Warwick B. Dunn2,3†, Garth J.S. Cooper2,3,4,5* & Natalie J. Gardiner1* 1 Faculty of Life Sciences, University of Manchester, UK 2 Centre for Advanced Discovery and Experimental Therapeutics (CADET), Central Manchester University Hospitals NHS Foundation Trust, Manchester Academic Health Sciences Centre, Manchester, UK 3 Centre for Endocrinology and Diabetes, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, UK 4 School of Biological Sciences, University of Auckland, New Zealand 5 Department of Pharmacology, Medical Sciences Division, University of Oxford, UK † Present address: School of Biosciences, University of Birmingham, UK *Joint corresponding authors: Natalie J. Gardiner and Garth J.S. Cooper Email: [email protected]; [email protected] Address: University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, United Kingdom Telephone: +44 161 275 5768; +44 161 701 0240 Word count: 4,490 Number of tables: 1, Number of figures: 6 Running title: Metabolic dysfunction in diabetic neuropathy 1 Diabetes Publish Ahead of Print, published online October 15, 2015 Diabetes Page 2 of 255 Abstract High glucose levels in the peripheral nervous system (PNS) have been implicated in the pathogenesis of diabetic neuropathy (DN). However our understanding of the molecular mechanisms which cause the marked distal pathology is incomplete. Here we performed a comprehensive, system-wide analysis of the PNS of a rodent model of DN. -

Targeting Tyro3 Ameliorates a Model of PGRN-Mutant FTLD-TDP Via Tau
ARTICLE DOI: 10.1038/s41467-018-02821-z OPEN Targeting Tyro3 ameliorates a model of PGRN- mutant FTLD-TDP via tau-mediated synaptic pathology Kyota Fujita1, Xigui Chen1, Hidenori Homma1, Kazuhiko Tagawa1, Mutsuki Amano 2, Ayumu Saito3, Seiya Imoto 4, Hiroyasu Akatsu5, Yoshio Hashizume6, Kozo Kaibuchi2, Satoru Miyano3 & Hitoshi Okazawa1 PGRN 1234567890():,; Mutations in the progranulin ( ) gene cause a tau pathology-negative and TDP43 pathology-positive form of frontotemporal lobar degeneration (FTLD-TDP). We generated a knock-in mouse harboring the R504X mutation (PGRN-KI). Phosphoproteomic analysis of this model revealed activation of signaling pathways connecting PKC and MAPK to tau prior to TDP43 aggregation and cognitive impairments, and identified PKCα as the kinase responsible for the early-stage tau phosphorylation at Ser203. Disinhibition of Gas6 binding to Tyro3 due to PGRN reduction results in activation of PKCα via PLCγ, inducing tau phosphorylation at Ser203, mislocalization of tau to dendritic spines, and spine loss. Administration of a PKC inhibitor, B-Raf inhibitor, or knockdown of molecules in the Gas6-Tyro3-tau axis rescues spine loss and cognitive impairment of PGRN-KI mice. Collectively, these results suggest that targeting of early-stage and aggregation-independent tau signaling represents a promising therapeutic strategy for this disease. 1 Department of Neuropathology, Medical Research Institute and Center for Brain Integration Research, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan. 2 Department of Cell Pharmacology, Graduate School of Medicine, Nagoya University, 65, Tsurumai, Showa, Nagoya, Aichi 466-8550, Japan. 3 Human Genome Center, Institute of Medical Science, The University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo 108- 8639, Japan. -

HNRPH1 (HNRNPH1) (NM 005520) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC201834 HNRPH1 (HNRNPH1) (NM_005520) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: HNRPH1 (HNRNPH1) (NM_005520) Human Tagged ORF Clone Tag: Myc-DDK Symbol: HNRNPH1 Synonyms: hnRNPH; HNRPH; HNRPH1 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 6 HNRPH1 (HNRNPH1) (NM_005520) Human Tagged ORF Clone – RC201834 ORF Nucleotide >RC201834 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGATGTTGGGCACGGAAGGTGGAGAGGGATTCGTGGTGAAGGTCCGGGGCTTGCCCTGGTCTTGCTCGG CCGATGAAGTGCAGAGGTTTTTTTCTGACTGCAAAATTCAAAATGGGGCTCAAGGTATTCGTTTCATCTA CACCAGAGAAGGCAGACCAAGTGGCGAGGCTTTTGTTGAACTTGAATCAGAAGATGAAGTCAAATTGGCC CTGAAAAAAGACAGAGAAACTATGGGACACAGATATGTTGAAGTATTCAAGTCAAACAACGTTGAAATGG ATTGGGTGTTGAAGCATACTGGTCCAAATAGTCCTGACACGGCCAATGATGGCTTTGTACGGCTTAGAGG ACTTCCCTTTGGATGTAGCAAGGAAGAAATTGTTCAGTTCTTCTCAGGGTTGGAAATCGTGCCAAATGGG ATAACATTGCCGGTGGACTTCCAGGGGAGGAGTACGGGGGAGGCCTTCGTGCAGTTTGCTTCACAGGAAA TAGCTGAAAAGGCTCTAAAGAAACACAAGGAAAGAATAGGGCACAGGTATATTGAAATCTTTAAGAGCAG TAGAGCTGAAGTTAGAACTCATTATGATCCACCACGAAAGCTTATGGCCATGCAGCGGCCAGGTCCTTAT -
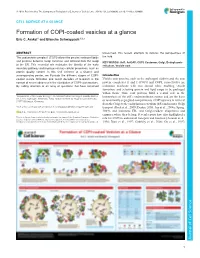
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Snapshot: the Splicing Regulatory Machinery Mathieu Gabut, Sidharth Chaudhry, and Benjamin J
192 Cell SnapShot: The Splicing Regulatory Machinery Mathieu Gabut, Sidharth Chaudhry, and Benjamin J. Blencowe 133 Banting and Best Department of Medical Research, University of Toronto, Toronto, ON M5S 3E1, Canada Expression in mouse , April4, 2008©2008Elsevier Inc. Low High Name Other Names Protein Domains Binding Sites Target Genes/Mouse Phenotypes/Disease Associations Amy Ceb Hip Hyp OB Eye SC BM Bo Ht SM Epd Kd Liv Lu Pan Pla Pro Sto Spl Thy Thd Te Ut Ov E6.5 E8.5 E10.5 SRp20 Sfrs3, X16 RRM, RS GCUCCUCUUC SRp20, CT/CGRP; −/− early embryonic lethal E3.5 9G8 Sfrs7 RRM, RS, C2HC Znf (GAC)n Tau, GnRH, 9G8 ASF/SF2 Sfrs1 RRM, RS RGAAGAAC HipK3, CaMKIIδ, HIV RNAs; −/− embryonic lethal, cond. KO cardiomyopathy SC35 Sfrs2 RRM, RS UGCUGUU AChE; −/− embryonic lethal, cond. KO deficient T-cell maturation, cardiomyopathy; LS SRp30c Sfrs9 RRM, RS CUGGAUU Glucocorticoid receptor SRp38 Fusip1, Nssr RRM, RS ACAAAGACAA CREB, type II and type XI collagens SRp40 Sfrs5, HRS RRM, RS AGGAGAAGGGA HipK3, PKCβ-II, Fibronectin SRp55 Sfrs6 RRM, RS GGCAGCACCUG cTnT, CD44 DOI 10.1016/j.cell.2008.03.010 SRp75 Sfrs4 RRM, RS GAAGGA FN1, E1A, CD45; overexpression enhances chondrogenic differentiation Tra2α Tra2a RRM, RS GAAARGARR GnRH; overexpression promotes RA-induced neural differentiation SR and SR-Related Proteins Tra2β Sfrs10 RRM, RS (GAA)n HipK3, SMN, Tau SRm160 Srrm1 RS, PWI AUGAAGAGGA CD44 SWAP Sfrs8 RS, SWAP ND SWAP, CD45, Tau; possible asthma susceptibility gene hnRNP A1 Hnrnpa1 RRM, RGG UAGGGA/U HipK3, SMN2, c-H-ras; rheumatoid arthritis, systemic lupus -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Genetic and Pharmacological Approaches to Preventing Neurodegeneration
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Genetic and Pharmacological Approaches to Preventing Neurodegeneration Marco Boccitto University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Neuroscience and Neurobiology Commons Recommended Citation Boccitto, Marco, "Genetic and Pharmacological Approaches to Preventing Neurodegeneration" (2012). Publicly Accessible Penn Dissertations. 494. https://repository.upenn.edu/edissertations/494 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/494 For more information, please contact [email protected]. Genetic and Pharmacological Approaches to Preventing Neurodegeneration Abstract The Insulin/Insulin-like Growth Factor 1 Signaling (IIS) pathway was first identified as a major modifier of aging in C.elegans. It has since become clear that the ability of this pathway to modify aging is phylogenetically conserved. Aging is a major risk factor for a variety of neurodegenerative diseases including the motor neuron disease, Amyotrophic Lateral Sclerosis (ALS). This raises the possibility that the IIS pathway might have therapeutic potential to modify the disease progression of ALS. In a C. elegans model of ALS we found that decreased IIS had a beneficial effect on ALS pathology in this model. This beneficial effect was dependent on activation of the transcription factor daf-16. To further validate IIS as a potential therapeutic target for treatment of ALS, manipulations of IIS in mammalian cells were investigated for neuroprotective activity. Genetic manipulations that increase the activity of the mammalian ortholog of daf-16, FOXO3, were found to be neuroprotective in a series of in vitro models of ALS toxicity.