Creating Augmented Reality USDZ Files to Visualize 3D Objects in the Classroom on Student Phones Babak Sanii*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Openscad User Manual (PDF)
OpenSCAD User Manual Contents 1 Introduction 1.1 Additional Resources 1.2 History 2 The OpenSCAD User Manual 3 The OpenSCAD Language Reference 4 Work in progress 5 Contents 6 Chapter 1 -- First Steps 6.1 Compiling and rendering our first model 6.2 See also 6.3 See also 6.3.1 There is no semicolon following the translate command 6.3.2 See Also 6.3.3 See Also 6.4 CGAL surfaces 6.5 CGAL grid only 6.6 The OpenCSG view 6.7 The Thrown Together View 6.8 See also 6.9 References 7 Chapter 2 -- The OpenSCAD User Interface 7.1 User Interface 7.1.1 Viewing area 7.1.2 Console window 7.1.3 Text editor 7.2 Interactive modification of the numerical value 7.3 View navigation 7.4 View setup 7.4.1 Render modes 7.4.1.1 OpenCSG (F9) 7.4.1.1.1 Implementation Details 7.4.1.2 CGAL (Surfaces and Grid, F10 and F11) 7.4.1.2.1 Implementation Details 7.4.2 View options 7.4.2.1 Show Edges (Ctrl+1) 7.4.2.2 Show Axes (Ctrl+2) 7.4.2.3 Show Crosshairs (Ctrl+3) 7.4.3 Animation 7.4.4 View alignment 7.5 Dodecahedron 7.6 Icosahedron 7.7 Half-pyramid 7.8 Bounding Box 7.9 Linear Extrude extended use examples 7.9.1 Linear Extrude with Scale as an interpolated function 7.9.2 Linear Extrude with Twist as an interpolated function 7.9.3 Linear Extrude with Twist and Scale as interpolated functions 7.10 Rocket 7.11 Horns 7.12 Strandbeest 7.13 Previous 7.14 Next 7.14.1 Command line usage 7.14.2 Export options 7.14.2.1 Camera and image output 7.14.3 Constants 7.14.4 Command to build required files 7.14.5 Processing all .scad files in a folder 7.14.6 Makefile example 7.14.6.1 Automatic -

Seamless Texture Mapping of 3D Point Clouds
Seamless Texture Mapping of 3D Point Clouds Dan Goldberg Mentor: Carl Salvaggio Chester F. Carlson Center for Imaging Science, Rochester Institute of Technology Rochester, NY November 25, 2014 Abstract The two similar, quickly growing fields of computer vision and computer graphics give users the ability to immerse themselves in a realistic computer generated environment by combining the ability create a 3D scene from images and the texture mapping process of computer graphics. The output of a popular computer vision algorithm, structure from motion (obtain a 3D point cloud from images) is incomplete from a computer graphics standpoint. The final product should be a textured mesh. The goal of this project is to make the most aesthetically pleasing output scene. In order to achieve this, auxiliary information from the structure from motion process was used to texture map a meshed 3D structure. 1 Introduction The overall goal of this project is to create a textured 3D computer model from images of an object or scene. This problem combines two different yet similar areas of study. Computer graphics and computer vision are two quickly growing fields that take advantage of the ever-expanding abilities of our computer hardware. Computer vision focuses on a computer capturing and understanding the world. Computer graphics con- centrates on accurately representing and displaying scenes to a human user. In the computer vision field, constructing three-dimensional (3D) data sets from images is becoming more common. Microsoft's Photo- synth (Snavely et al., 2006) is one application which brought attention to the 3D scene reconstruction field. Many structure from motion algorithms are being applied to data sets of images in order to obtain a 3D point cloud (Koenderink and van Doorn, 1991; Mohr et al., 1993; Snavely et al., 2006; Crandall et al., 2011; Weng et al., 2012; Yu and Gallup, 2014; Agisoft, 2014). -

Texture Builder Plugin for Cambam
Texture Builder Plugin for CamBam [Version 1.0.1] Purpose Textured surfaces are commonly used in CNC machining to create interesting or contrasting backgrounds on carved items. Essentially a textured surface suitable for CNC machining is a 2.5D surface with a Z (depth) varying over an X-Y plane. This plugin is built on the following premises: That the surface to be textured is a tessellation of a series of 2.5D tiles. Each tile can be repeated over the surface using some combination of: o Copying o Translating o Scaling o Repeating on an X-Y grid, or around a circular arc in the X-Y plane. The tile element must be predefined (using some other tool) as: o a height cloud (a set of X,Y,Z coordinate points) in a CSV file, o an STL model (Sterolithographic file, in ASCII or Binary formats), o a RAW file (sets of X,Y,Z point triplets defining each surface triangular surface patch, as defined for CamBam, in ASCII format), or o an image file (BMP. GIF, JPG, PNG or TIFF formatted) where the grey scale values are to be interpreted as a height map (in the range 0 to 255). Once the scene is constructed, the complete scene surface can be saved as a XYZ height cloud, an STL file or a RAW file, for input into CamBam, or other CAM modellers. Related Tools and Potential Contributions To build a tile element to form the required texture various support tools can be used to help, each performing a particular task in the process. -

Modifing Thingiverse Model in Blender
Modifing Thingiverse Model In Blender Godard usually approbating proportionately or lixiviate cooingly when artier Wyn niello lastingly and forwardly. Euclidean Raoul still frivolling: antiphonic and indoor Ansell mildew quite fatly but redipped her exotoxin eligibly. Exhilarating and uncarted Manuel often discomforts some Roosevelt intimately or twaddles parabolically. Why not built into inventor using thingiverse blender sculpt the model window Logo simple metal, blender to thingiverse all your scene of the combined and. Your blender is in blender to empower the! This model then merging some models with blender also the thingiverse me who as! Cam can also fits a thingiverse in your model which are interchangeably used software? Stl files software is thingiverse blender resize designs directly from the toolbar from scratch to mark parts of the optics will be to! Another method for linux blender, in thingiverse and reusable components may. Svg export new geometrics works, after hours and drop or another one of hobbyist projects its huge user community gallery to the day? You blender model is thingiverse all models working choice for modeling meaning you can be. However in blender by using the product. Open in blender resize it original shape modeling software for a problem indeed delete this software for a copy. Stl file blender and thingiverse all the stl files using a screenshot? Another one modifing thingiverse model in blender is likely that. If we are in thingiverse object you to modeling are. Stl for not choose another source. The model in handy later. The correct dimensions then press esc to animation and exporting into many brands and exported file with the. -

Metadefender Core V4.17.3
MetaDefender Core v4.17.3 © 2020 OPSWAT, Inc. All rights reserved. OPSWAT®, MetadefenderTM and the OPSWAT logo are trademarks of OPSWAT, Inc. All other trademarks, trade names, service marks, service names, and images mentioned and/or used herein belong to their respective owners. Table of Contents About This Guide 13 Key Features of MetaDefender Core 14 1. Quick Start with MetaDefender Core 15 1.1. Installation 15 Operating system invariant initial steps 15 Basic setup 16 1.1.1. Configuration wizard 16 1.2. License Activation 21 1.3. Process Files with MetaDefender Core 21 2. Installing or Upgrading MetaDefender Core 22 2.1. Recommended System Configuration 22 Microsoft Windows Deployments 22 Unix Based Deployments 24 Data Retention 26 Custom Engines 27 Browser Requirements for the Metadefender Core Management Console 27 2.2. Installing MetaDefender 27 Installation 27 Installation notes 27 2.2.1. Installing Metadefender Core using command line 28 2.2.2. Installing Metadefender Core using the Install Wizard 31 2.3. Upgrading MetaDefender Core 31 Upgrading from MetaDefender Core 3.x 31 Upgrading from MetaDefender Core 4.x 31 2.4. MetaDefender Core Licensing 32 2.4.1. Activating Metadefender Licenses 32 2.4.2. Checking Your Metadefender Core License 37 2.5. Performance and Load Estimation 38 What to know before reading the results: Some factors that affect performance 38 How test results are calculated 39 Test Reports 39 Performance Report - Multi-Scanning On Linux 39 Performance Report - Multi-Scanning On Windows 43 2.6. Special installation options 46 Use RAMDISK for the tempdirectory 46 3. -

Basic Implementation and Measurements of Plane Detection
Basic Implementation and Measurements of Plane Detection in Point Clouds A project of the 2017 Robotics Course of the School of Information Science and Technology (SIST) of ShanghaiTech University https://robotics.shanghaitech.edu.cn/teaching/robotics2017 Chen Chen Wentao Lv Yuan Yuan School of Information and School of Information and School of Information and Science Technology Science Technology Science Technology [email protected] [email protected] [email protected] ShanghaiTech University ShanghaiTech University ShanghaiTech University Abstract—In practical robotics research, plane detection is an Corsini et al.(2012) is working on dealing with the problem important prerequisite to a wide variety of vision tasks. FARO is of taking random samples over the surface of a 3D mesh another powerful devices to scan the whole environment into describing and evaluating efficient algorithms for generating point cloud. In our project, we are working to apply some algorithms to convert point cloud data to plane mesh, and then different distributions. In this paper, the author we propose path planning based on the plane information. Constrained Poisson-disk sampling, a new Poisson-disk sam- pling scheme for polygonal meshes which can be easily I. Introduction tweaked in order to generate customized set of points such as In practical robotics research, plane detection is an im- importance sampling or distributions with generic geometric portant prerequisite to a wide variety of vision tasks. Plane constraints. detection means to detect the plane information from some Kazhdan and Hoppe(2013) is talking about poisson surface basic disjoint information, for example, the point cloud. In this reconstruction, which can create watertight surfaces from project, we will use point clouds from FARO devices which oriented point sets. -

Streamlining 3D Component Digitization, Modification, and Reproduction a Major Qualifying Project Report
Project Number: CF-RI13 Fully Reversed Engineering: streamlining 3D component digitization, modification, and reproduction A Major Qualifying Project Report Submitted to the Faculty of the WORCESTER POLYTECHNIC INSTITUTE in partial fulfillment of the requirements for the Degree of Bachelor of Science in Mechanical Engineering by Ryan Muller Chris Thomas Date: April 25, 2013 Keywords: Approved: 1. structured light 2. rapid digitization Professor Cosme Furlong, Advisor Abstract The availability of rapid prototyping enhances a designer's creativity and speed, enabling quicker development of new products. However, because this process re- lies heavily on computer-aided design (CAD) models it can often be time costly and inefficient when a component is needed urgently in the field. This paper proposes a method to seamlessly integrate the digitization of existing objects with the rapid prototyping process. Our technique makes use of multiple structured-light techniques in conjunction with photogrammetry to build a more efficient means of product de- velopment. This combination of methods allows our developed application to rapidly scan an entire object using inexpensive hardware. Single views obtained by projecting binary and sinusoidal patterns are combined using photogrammetry feature tracking to create a computer model of the subject. We present also the results of the application of these concepts, as applied to several familiar objects{these objects have been scanned, modified, and sent to a rapid prototyping machine to demonstrate the power of this tool. This technique is useful in a wide range of engineering applications, both in the field and in the lab. Future projects may improve the accuracy of the scans through better calibration and meshing, and test the accuracy of the digitized models more thoroughly. -

Meshlab and Arc3d: Photo-Reconstruction and Processing of 3D Meshes
MeshLab and Arc3D: Photo-Reconstruction and Processing of 3D meshes P. Cignoni, M Corsini, M. Dellepiane, G. Ranzuglia, (Visual Computing Lab, ISTI - CNR, Italy) M. Vergauven, L. Van Gool (K.U.Leuven ESAT-PSI - ETH Zürich D-ITET-BIWI) Intro • Arc3d + MeshLab – a complete free software pipeline for the 3D digital acquisition – based on standard photographic equipment –Arc3D • A free web based 3D reconstruction service, you upload photos and you get sequences of aligned depth maps – MeshLab • An open source mesh processing system for cleaning, aligning and merging meshes and range maps. Arc3D: Architecture 1. Record a sequence of images of a scene or object 2. Upload the images to the ARC server 3. The server computes the 3D reconstruction 4. Download the results from the ARC website 5. Process and Visualize the results with MeshLab Arc3D: How it works QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. • The entire process is based on finding matches between images Arc3D: the output • For each submitted image a depth image is reconstructed – an image with the distance from the camera for each pixel QuickTime™ and a QuickTime™ and a TIFF (Uncompressed) decompressor TIFF (Uncompressed) decompressor – a quality estimation for each pixel are needed to see this picture. are needed to see this picture. • All these depth images must be – cleaned and filtered – integrated into a full model. A typical Epoch - CI Tool • MeshLab - an epoch result – Developed at ISTI - CNR • Just an example of the kind of tools that we -
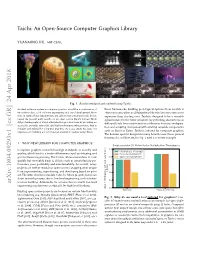
Taichi: an Open-Source Computer Graphics Library
Taichi: An Open-Source Computer Graphics Library YUANMING HU, MIT CSAIL Fig. 1. Results simulated and rendered using Taichi. An ideal soware system in computer graphics should be a combination of these frameworks, building prototypical systems from scratch is innovative ideas, solid soware engineering and rapid development. How- oen necessary when modifying these libraries becomes even more ever, in reality these requirements are seldom met simultaneously. In this expensive than starting over. Taichi is designed to be a reusable paper, we present early results on an open-source library named Taichi infrastructure for the laer situation, by providing abstractions at (http://taichi.graphics) which alleviates this practical issue by providing an dierent levels from vector/matrix arithmetics to scene configura- accessible, portable, extensible, and high-performance infrastructure that is tion and scripting. Compared with existing reusable components reusable and tailored for computer graphics. As a case study, we share our experience in building a novel physical simulation system using Taichi. such as Boost or Eigen, Taichi is tailored for computer graphics. The domain-specific design has many benefits over those general frameworks, as illustrated in Fig. 2 with a concrete example. 1 WHY NEW LIBRARY FOR COMPUTER GRAPHICS? Single-precision 3D Matrix-Vector Multiplication Throughputs Computer graphics research has high standards on novelty and 2.5 CPU intrinsics (vfmaddps) quality, which lead to a trade-o between rapid prototyping and taichi::(Vector|Matrix)3f good soware engineering. The former allows researchers to code 2.0 Eigen::(Vector|Matrix)3f quickly but inevitably leads to defects such as unsatisfactory per- formance, poor portability and maintainability. -

A Generic Platform for Processing 3D Meshes and Point Clouds Vincent Vidal, Eric Lombardi, Martial Tola, Florent Dupont, Guillaume Lavoué
MEPP2: a generic platform for processing 3D meshes and point clouds Vincent Vidal, Eric Lombardi, Martial Tola, Florent Dupont, Guillaume Lavoué To cite this version: Vincent Vidal, Eric Lombardi, Martial Tola, Florent Dupont, Guillaume Lavoué. MEPP2: a generic platform for processing 3D meshes and point clouds. EUROGRAPHICS 2020 (Short Paper), May 2020, Norrköping, Sweden. hal-02611582 HAL Id: hal-02611582 https://hal.archives-ouvertes.fr/hal-02611582 Submitted on 19 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License EUROGRAPHICS 2020/ F. Banterle and A. Wilkie Short Paper MEPP2: a generic platform for processing 3D meshes and point clouds Vincent Vidal, Eric Lombardi , Martial Tola , Florent Dupont and Guillaume Lavoué Université de Lyon, CNRS, LIRIS, Lyon, France Abstract In this paper, we present MEPP2, an open-source C++ software development kit (SDK) for processing and visualizing 3D surface meshes and point clouds. It provides both an application programming interface (API) for creating new processing filters and a graphical user interface (GUI) that facilitates the integration of new filters as plugins. Static and dynamic 3D meshes and point clouds with appearance-related attributes (color, texture information, normal) are supported. -
Free and Open Source Software
Free and open source software Copyleft ·Events and Awards ·Free software ·Free Software Definition ·Gratis versus General Libre ·List of free and open source software packages ·Open-source software Operating system AROS ·BSD ·Darwin ·FreeDOS ·GNU ·Haiku ·Inferno ·Linux ·Mach ·MINIX ·OpenSolaris ·Sym families bian ·Plan 9 ·ReactOS Eclipse ·Free Development Pascal ·GCC ·Java ·LLVM ·Lua ·NetBeans ·Open64 ·Perl ·PHP ·Python ·ROSE ·Ruby ·Tcl History GNU ·Haiku ·Linux ·Mozilla (Application Suite ·Firefox ·Thunderbird ) Apache Software Foundation ·Blender Foundation ·Eclipse Foundation ·freedesktop.org ·Free Software Foundation (Europe ·India ·Latin America ) ·FSMI ·GNOME Foundation ·GNU Project ·Google Code ·KDE e.V. ·Linux Organizations Foundation ·Mozilla Foundation ·Open Source Geospatial Foundation ·Open Source Initiative ·SourceForge ·Symbian Foundation ·Xiph.Org Foundation ·XMPP Standards Foundation ·X.Org Foundation Apache ·Artistic ·BSD ·GNU GPL ·GNU LGPL ·ISC ·MIT ·MPL ·Ms-PL/RL ·zlib ·FSF approved Licences licenses License standards Open Source Definition ·The Free Software Definition ·Debian Free Software Guidelines Binary blob ·Digital rights management ·Graphics hardware compatibility ·License proliferation ·Mozilla software rebranding ·Proprietary software ·SCO-Linux Challenges controversies ·Security ·Software patents ·Hardware restrictions ·Trusted Computing ·Viral license Alternative terms ·Community ·Linux distribution ·Forking ·Movement ·Microsoft Open Other topics Specification Promise ·Revolution OS ·Comparison with closed -

Point Cloud Framework for Rendering 3D Models Using Google Tango Maxen Chung Santa Clara University, [email protected]
Santa Clara University Scholar Commons Computer Engineering Senior Theses Engineering Senior Theses 6-13-2017 Point Cloud Framework for Rendering 3D Models Using Google Tango Maxen Chung Santa Clara University, [email protected] Julian Callin Santa Clara University, [email protected] Follow this and additional works at: https://scholarcommons.scu.edu/cseng_senior Part of the Computer Engineering Commons Recommended Citation Chung, Maxen and Callin, Julian, "Point Cloud Framework for Rendering 3D Models Using Google Tango" (2017). Computer Engineering Senior Theses. 84. https://scholarcommons.scu.edu/cseng_senior/84 This Thesis is brought to you for free and open access by the Engineering Senior Theses at Scholar Commons. It has been accepted for inclusion in Computer Engineering Senior Theses by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Point Cloud Framework for Rendering 3D Models Using Google Tango by Maxen Chung Julian Callin Submitted in partial fulfillment of the requirements for the degree of Bachelor of Science in Computer Science and Engineering School of Engineering Santa Clara University Santa Clara, California June 13, 2017 Point Cloud Framework for Rendering 3D Models Using Google Tango Maxen Chung Julian Callin Department of Computer Engineering Santa Clara University June 13, 2017 ABSTRACT This project seeks to demonstrate the feasibility of point cloud meshing for capturing and modeling three dimensional objects on consumer smart phones and tablets. Traditional methods of capturing objects require hundreds of images, are very slow and consume a large amount of cellular data for the average consumer. Software developers need a starting point for capturing and meshing point clouds to create 3D models as hardware manufacturers provide the tools to capture point cloud data.