The Role of N2A and N2B Titin Isoforms in Muscle Development By
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Muscular System
THE MUSCULAR SYSTEM COMPILED BY HOWIE BAUM 1 Muscles make up the bulk of the body and account for 1/3 of its weight.!! Blood vessels and nerves run to every muscle, helping control and regulate each muscle’s function. The muscular system creates body heat and also moves the: Bones of the Skeletal system Food through Digestive system Blood through the Circulatory system Fluids through the Excretory system MUSCLE TISSUE The body has 3 main types of muscle tissue 1) Skeletal, 2) Smooth, and 3) Cardiac SKELETAL MUSCLE SMOOTH MUSCLE CARDIAC MUSCLE Skeletal muscles attach to and move bones by contracting and relaxing in response to voluntary messages from the nervous system. Skeletal muscle tissue is composed of long cells called muscle fibers that have a striated appearance. Muscle fibers are organized into bundles supplied by blood vessels and innervated by motor neurons. Muscle structure Skeletal (striated or voluntary) muscle consists of densely packed groups of hugely elongated cells known as myofibers. These are grouped into bundles (fascicles). A typical myofiber is 2–3 centimeters ( 3/4–1 1/5 in) long and 0.05millimeters (1/500 inch) in diameter and is composed of narrower structures – myofibrils. These contain thick and thin myofilaments made up mainly of the proteins actin and myosin. Numerous capillaries keep the muscle supplied with the oxygen and glucose needed to fuel contraction. Skeletal Muscles • Skeletal muscles attach to bones by tendons (connective tissue) and enable movement. • Skeletal muscles are mostly voluntary Feel the back of your ankle to feel your Achilles tendon - the largest tendon in your body. -

(7E) Powerpoint Lecture Outline Chapter 8: Control of Movement
Carlson (7e) PowerPoint Lecture Outline Chapter 8: Control of Movement This multimedia product and its contents are protected under copyright law. The following are prohibited by law: •any public performance or display, including transmission of any image over a network; •preparation of any derivative work, including extraction, in whole or in part, of any images; •any rental, lease, or lending of the program. Copyright 2001 by Allyn & Bacon Skeletal Muscle n Movements of our body are accomplished by contraction of the skeletal muscles l Flexion: contraction of a flexor muscle draws in a limb l Extension: contraction of extensor muscle n Skeletal muscle fibers have a striated appearance n Skeletal muscle is composed of two fiber types: l Extrafusal: innervated by alpha-motoneurons from the spinal cord: exert force l Intrafusal: sensory fibers that detect stretch of the muscle u Afferent fibers: report length of intrafusal: when stretched, the fibers stimulate the alpha-neuron that innervates the muscle fiber: maintains muscle tone u Efferent fibers: contraction adjusts sensitivity of afferent fibers. 8.2 Copyright 2001 by Allyn & Bacon Skeletal Muscle Anatomy n Each muscle fiber consists of a bundle of myofibrils l Each myofibril is made up of overlapping strands of actin and myosin l During a muscle twitch, the myosin filaments move relative to the actin filaments, thereby shortening the muscle fiber 8.3 Copyright 2001 by Allyn & Bacon Neuromuscular Junction n The neuromuscular junction is the synapse formed between an alpha motor neuron -

Evidence for the Nonmuscle Nature of The" Myofibroblast" of Granulation Tissue and Hypertropic Scar. an Immunofluorescence Study
American Journal of Pathology, Vol. 130, No. 2, February 1988 Copyright i American Association of Pathologists Evidencefor the Nonmuscle Nature ofthe "Myofibroblast" ofGranulation Tissue and Hypertropic Scar An Immunofluorescence Study ROBERT J. EDDY, BSc, JANE A. PETRO, MD, From the Department ofAnatomy, the Department ofSurgery, and JAMES J. TOMASEK, PhD Division ofPlastic and Reconstructive Surgery, and the Department of Orthopaedic Surgery, New York Medical College, Valhalla, New York Contraction is an important phenomenon in wound cell-matrix attachment in smooth muscle and non- repair and hypertrophic scarring. Studies indicate that muscle cells, respectively. Myofibroblasts can be iden- wound contraction involves a specialized cell known as tified by their intense staining of actin bundles with the myofibroblast, which has morphologic character- either anti-actin antibody or NBD-phallacidin. Myofi- istics of both smooth muscle and fibroblastic cells. In broblasts in all tissues stained for nonmuscle but not order to better characterize the myofibroblast, the au- smooth muscle myosin. In addition, nonmuscle myo- thors have examined its cytoskeleton and surrounding sin was localized as intracellular fibrils, which suggests extracellular matrix (ECM) in human burn granula- their similarity to stress fibers in cultured fibroblasts. tion tissue, human hypertrophic scar, and rat granula- The ECM around myofibroblasts stains intensely for tion tissue by indirect immunofluorescence. Primary fibronectin but lacks laminin, which suggests that a antibodies used in this study were directed against 1) true basal lamina is not present. The immunocytoche- smooth muscle myosin and 2) nonmuscle myosin, mical findings suggest that the myofibroblast is a spe- components ofthe cytoskeleton in smooth muscle and cialized nonmuscle type of cell, not a smooth muscle nonmuscle cells, respectively, and 3) laminin and 4) cell. -

Nomina Histologica Veterinaria, First Edition
NOMINA HISTOLOGICA VETERINARIA Submitted by the International Committee on Veterinary Histological Nomenclature (ICVHN) to the World Association of Veterinary Anatomists Published on the website of the World Association of Veterinary Anatomists www.wava-amav.org 2017 CONTENTS Introduction i Principles of term construction in N.H.V. iii Cytologia – Cytology 1 Textus epithelialis – Epithelial tissue 10 Textus connectivus – Connective tissue 13 Sanguis et Lympha – Blood and Lymph 17 Textus muscularis – Muscle tissue 19 Textus nervosus – Nerve tissue 20 Splanchnologia – Viscera 23 Systema digestorium – Digestive system 24 Systema respiratorium – Respiratory system 32 Systema urinarium – Urinary system 35 Organa genitalia masculina – Male genital system 38 Organa genitalia feminina – Female genital system 42 Systema endocrinum – Endocrine system 45 Systema cardiovasculare et lymphaticum [Angiologia] – Cardiovascular and lymphatic system 47 Systema nervosum – Nervous system 52 Receptores sensorii et Organa sensuum – Sensory receptors and Sense organs 58 Integumentum – Integument 64 INTRODUCTION The preparations leading to the publication of the present first edition of the Nomina Histologica Veterinaria has a long history spanning more than 50 years. Under the auspices of the World Association of Veterinary Anatomists (W.A.V.A.), the International Committee on Veterinary Anatomical Nomenclature (I.C.V.A.N.) appointed in Giessen, 1965, a Subcommittee on Histology and Embryology which started a working relation with the Subcommittee on Histology of the former International Anatomical Nomenclature Committee. In Mexico City, 1971, this Subcommittee presented a document entitled Nomina Histologica Veterinaria: A Working Draft as a basis for the continued work of the newly-appointed Subcommittee on Histological Nomenclature. This resulted in the editing of the Nomina Histologica Veterinaria: A Working Draft II (Toulouse, 1974), followed by preparations for publication of a Nomina Histologica Veterinaria. -

Regulation of Titin-Based Cardiac Stiffness by Unfolded Domain Oxidation (Undox)
Regulation of titin-based cardiac stiffness by unfolded domain oxidation (UnDOx) Christine M. Loeschera,1, Martin Breitkreuzb,1, Yong Lia, Alexander Nickelc, Andreas Ungera, Alexander Dietld, Andreas Schmidte, Belal A. Mohamedf, Sebastian Kötterg, Joachim P. Schmitth, Marcus Krügere,i, Martina Krügerg, Karl Toischerf, Christoph Maackc, Lars I. Leichertj, Nazha Hamdanib, and Wolfgang A. Linkea,2 aInstitute of Physiology II, University of Munster, 48149 Munster, Germany; bInstitute of Physiology, Ruhr University Bochum, 44801 Bochum, Germany; cComprehensive Heart Failure Center Wuerzburg, University Clinic Wuerzburg, 97078 Wuerzburg, Germany; dDepartment of Internal Medicine II, University Hospital Regensburg, 93053 Regensburg, Germany; eInstitute for Genetics, University of Cologne, 50931 Cologne, Germany; fDepartment of Cardiology and Pneumology, University Medicine Goettingen, 37075 Goettingen, Germany; gDepartment of Cardiovascular Physiology, Heinrich Heine University, 40225 Düsseldorf, Germany; hDepartment of Pharmacology and Clinical Pharmacology, Heinrich Heine University, 40225 Düsseldorf, Germany; iCenter for Molecular Medicine and Excellence Cluster "Cellular Stress Responses in Aging-Associated Diseases" (CECAD), University of Cologne, 50931 Cologne, Germany; and jInstitute for Biochemistry and Pathobiochemistry, Ruhr University Bochum, 44801 Bochum, Germany Edited by Jonathan Seidman, Harvard University, Boston, MA, and approved August 12, 2020 (received for review March 14, 2020) The relationship between oxidative stress and -

Enhanced Upregulation of Smooth Muscle Related Transcripts by Tgfb2 in Asthmatic (Myo) Fibroblasts J Wicks, H M Haitchi, S T Holgate, D E Davies, R M Powell
313 ASTHMA Thorax: first published as 10.1136/thx.2005.050005 on 31 January 2006. Downloaded from Enhanced upregulation of smooth muscle related transcripts by TGFb2 in asthmatic (myo) fibroblasts J Wicks, H M Haitchi, S T Holgate, D E Davies, R M Powell ............................................................................................................................... Thorax 2006;61:313–319. doi: 10.1136/thx.2005.050005 Background: Transforming growth factor beta (TGFb) upregulates a number of smooth muscle specific genes in (myo)fibroblasts. As asthma is characterised by an increase in airway smooth muscle, we postulated that TGFb2 favours differentiation of asthmatic (myo)fibroblasts towards a smooth muscle phenotype. Methods: Primary fibroblasts were grown from bronchial biopsy specimens from normal (n = 6) and asthmatic (n = 7) donors and treated with TGFb2 to induce myofibroblast differentiation. The most stable genes for normalisation were identified using RT-qPCR and the geNorm software applied to a panel of 12 See end of article for authors’ affiliations ‘‘housekeeping’’ genes. Expression of a-smooth muscle actin (aSMA), heavy chain myosin (HCM), ....................... calponin 1 (CPN 1), desmin, and c-actin were measured by RT-qPCR. Protein expression was assessed by immunocytochemistry and western blotting. Correspondence to: Dr R M Powell, The Brooke Results: Phospholipase A2 and ubiquitin C were identified as the most stably expressed and practically Laboratory, Level F, South useful genes for normalisation of gene expression during myofibroblast differentiation. TGFb2 induced Block, Southampton mRNA expression for all five smooth muscle related transcripts; aSMA, HCM and CPN 1 protein were also General Hospital, Southampton SO16 6YD, increased but desmin protein was not detectable. Although there was no difference in basal expression, UK; [email protected] HCM, CPN 1 and desmin were induced to a significantly greater extent in asthmatic fibroblasts than in those from normal controls (p = 0.041 and 0.011, respectively). -

Postmortem Changes in the Myofibrillar and Other Cytoskeletal Proteins in Muscle
BIOCHEMISTRY - IMPACT ON MEAT TENDERNESS Postmortem Changes in the Myofibrillar and Other C'oskeletal Proteins in Muscle RICHARD M. ROBSON*, ELISABETH HUFF-LONERGAN', FREDERICK C. PARRISH, JR., CHIUNG-YING HO, MARVIN H. STROMER, TED W. HUIATT, ROBERT M. BELLIN and SUZANNE W. SERNETT introduction filaments (titin), and integral Z-line region (a-actinin, Cap Z), as well as proteins of the intermediate filaments (desmin, The cytoskeleton of "typical" vertebrate cells contains paranemin, and synemin), Z-line periphery (filamin) and three protein filament systems, namely the -7-nm diameter costameres underlying the cell membrane (filamin, actin-containing microfilaments, the -1 0-nm diameter in- dystrophin, talin, and vinculin) are listed along with an esti- termediate filaments (IFs), and the -23-nm diameter tubu- mate of their abundance, approximate molecular weights, lin-containing microtubules (Robson, 1989, 1995; Robson and number of subunits per molecule. Because the myofibrils et al., 1991 ).The contractile myofibrils, which are by far the are the overwhelming components of the skeletal muscle cell major components of developed skeletal muscle cells and cytoskeleton, the approximate percentages of the cytoskel- are responsible for most of the desirable qualities of muscle eton listed for the myofibrillar proteins (e.g., myosin, actin, foods (Robson et al., 1981,1984, 1991 1, can be considered tropomyosin, a-actinin, etc.) also would represent their ap- the highly expanded corollary of the microfilament system proximate percentages of total myofibrillar protein. of non-muscle cells. The myofibrils, IFs, cell membrane skel- eton (complex protein-lattice subjacent to the sarcolemma), Some Important Characteristics, Possible and attachment sites connecting these elements will be con- Roles, and Postmortem Changes of Key sidered as comprising the muscle cell cytoskeleton in this Cytoskeletal Proteins review. -

Α-Actinin/Titin Interaction: a Dynamic and Mechanically Stable Cluster of Bonds in the Muscle Z-Disk
α-Actinin/titin interaction: A dynamic and mechanically stable cluster of bonds in the muscle Z-disk Marco Grisona, Ulrich Merkela, Julius Kostanb, Kristina Djinovic-Carugob,c, and Matthias Riefa,d,1 aPhysik Department E22, Technische Universität München, 85748 Garching, Germany; bDepartment of Structural and Computational Biology, Max F. Perutz Laboratories, University of Vienna, A-1030 Vienna, Austria; cDepartment of Biochemistry, Faculty of Chemistry and Chemical Technology, University of Ljubljana, SI-1000 Ljubljana, Slovenia; and dMunich Center for Integrated Protein Science, 81377 Munich, Germany Edited by James A. Spudich, Stanford University School of Medicine, Stanford, CA, and approved December 16, 2016 (received for review August 2, 2016) Stable anchoring of titin within the muscle Z-disk is essential for In humans, the isoforms of titin exhibit four to seven Z-repeats preserving muscle integrity during passive stretching. One of the (15, 16, 20). The structure of the EF3-4 hands complex with titin main candidates for anchoring titin in the Z-disk is the actin cross- Z-repeat 7 shows the bound Z-repeat in an α-helical confor- linker α-actinin. The calmodulin-like domain of α-actinin binds to mation (21). In solution assays, binding affinities of various the Z-repeats of titin. However, the mechanical and kinetic prop- Z-repeats to EF3-4 were determined to lie in the micromolar erties of this important interaction are still unknown. Here, we use range (22). Micromolar affinity points only to a moderately a dual-beam optical tweezers assay to study the mechanics of this stable interaction, the kinetics of which are unknown. -
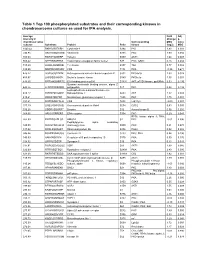
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

"Slow" and "Fast" Muscle Fibers
"Slow" and "Fast" Muscle Fibers ERNEST GUTMANN Department of Physiology and Pathophysiology of the Neuromuscular System, Institute of Physiology, Czechoslovak Academy of Sciences, Prague The great variety of structure and depolarization, but the slow fibers function of different muscles reflects of the frog show a sustained con apparently the process of adapta tracture during the whole period of tion to different functional de reduced membrane potential (Kuf mands, but it has remained a source fter, 1946; Fleckenstein, 1955). of confusion, especially if an at Thus two distinct fiber types, i.e., tempt is made to find a common fast (twitch) and slow (tonic) fi principle of order for this variety. bers do exist in frogs and toads and The terms "fast" and "slow" mus related differences have been cle fibers in mammals are used in described for the membrane poten reference to their faster or slower tials (Kuffier and Vaughan Wil contraction times. Both these mus liams, 1953; Kiessling, 1960), ar cles, e.g., the fast M. Extensor digi rangement of muscle fibrils torum (E.D.L.) and the slow M. ( "Fibrillen" or "Felderstruktur"), soleus of the rat are "twitch" mus pattern of filaments and structure cles, i.e., they react with propagated of the sarcoplasmatic reticulum action potentials to nerve stimula (Kruger, 1952 ; Gray, 1958 ; tion. Contrary to this, the slow Peachey, 1965), and type (focal or tonic muscle fibers of the frog re multiple) of innervation. spond with local non-propagated Kruger (1952) claimed that some depolarizations, activating contrac mammalian muscles consist en tures (Tasaki and Mizutani, 1943; tirely of fibers with "Felderstruk Kuffier and Gerard, 1947; Kuffier tur" and that the same distinction and Vaughan Williams, 1953). -

The Myofibroblast in Wound Healing and Fibrosis
F1000Research 2016, 5(F1000 Faculty Rev):752 Last updated: 17 JUL 2019 REVIEW The myofibroblast in wound healing and fibrosis: answered and unanswered questions [version 1; peer review: 2 approved] Marie-Luce Bochaton-Piallat1, Giulio Gabbiani1, Boris Hinz2 1Department of Pathology and Immunology, Faculty of Medicine, University of Geneva, Geneva, Switzerland 2Laboratory of Tissue Repair and Regeneration, Matrix Dynamics Group, Faculty of Dentistry, University of Toronto, Toronto, Canada First published: 26 Apr 2016, 5(F1000 Faculty Rev):752 ( Open Peer Review v1 https://doi.org/10.12688/f1000research.8190.1) Latest published: 26 Apr 2016, 5(F1000 Faculty Rev):752 ( https://doi.org/10.12688/f1000research.8190.1) Reviewer Status Abstract Invited Reviewers The discovery of the myofibroblast has allowed definition of the cell 1 2 responsible for wound contraction and for the development of fibrotic changes. This review summarizes the main features of the myofibroblast version 1 and the mechanisms of myofibroblast generation. Myofibroblasts originate published from a variety of cells according to the organ and the type of lesion. The 26 Apr 2016 mechanisms of myofibroblast contraction, which appear clearly different to those of smooth muscle cell contraction, are described. Finally, we summarize the possible strategies in order to reduce myofibroblast F1000 Faculty Reviews are written by members of activities and thus influence several pathologies, such as hypertrophic the prestigious F1000 Faculty. They are scars and organ fibrosis. commissioned and are peer reviewed before Keywords publication to ensure that the final, published version Myofibroblast , mechanotransduction , myofibroblast generation , is comprehensive and accessible. The reviewers myofibroblast contraction , hypertrophic scars , organ fibroses who approved the final version are listed with their names and affiliations. -

Association of Titin and Myosin Heavy Chain in Developing Skeletal Muscle (Myogenesis/Cytoskeleton/Assembly in Vvo) W
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 74%-7500, August 1992 Cell Biology Association of titin and myosin heavy chain in developing skeletal muscle (myogenesis/cytoskeleton/assembly in vvo) W. B. ISAACS*, I. S. KIM, A. STRUVE, AND A. B. FULTONt Department of Biochemistry, University of Iowa, Iowa City, IA 52242 Communicated by Sheldon Penman, May 22, 1992 ABSTRACT To understand molecular interactions that deficient medium (10). After labeling, cultures either were organize developing myoflbrils, we examined the biosynthesis extracted immediately or were chased by adding complete and interaction of titin and myosin heavy chain in cultures of medium supplemented with 2 mM unlabeled methionine and developing muscle. Use of pulse-labeling, immunoprecipita- incubating at 370C for various times before extraction. Ex- tion, and a reversible cross-linking procedure demonstrates tractions used 0.5% Triton X-100 in extraction buffer (100 that within minutes of synthesis, titin and myosin heavy chain mM KCI/10 mM Pipes, pH 6.8/300 mM sucrose/2 mM can be chemically cross-linked into very large, detergent- MgCI2/1 mM EGTA) containing protease inhibitors (1 mM resistant complexes retaining many features of intact myo- phenylmethylsulfonyl fluoride and 100 mM leupeptin; ref. tubes. These complexes, predominantly of titin and myosin, 11). occur very early in myofibrillogenesis as well as later. These Immunoprecipitation. Titin was precipitated by using a data suggest that synthesis and assembly oftitin and myosin are mouse monoclonal antibody (mAb), AMF-1, as described temporally and spatially coordinated in nascent myofibrils and (10). Muscle-specific myosin heavy chain (hereafter myosin) support the hypothesis that titin molecules help to organize was precipitated with mAb MF-20 (12), a gift ofD.