The Rise and Fall of Kin Structure in the Neotropical, Family-Living Buff-Breasted Wren (Cantorchilus Leucotis) in Gamboa, Panama
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Best of Costa Rica March 19–31, 2019
THE BEST OF COSTA RICA MARCH 19–31, 2019 Buffy-crowned Wood-Partridge © David Ascanio LEADERS: DAVID ASCANIO & MAURICIO CHINCHILLA LIST COMPILED BY: DAVID ASCANIO VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM THE BEST OF COSTA RICA March 19–31, 2019 By David Ascanio Photo album: https://www.flickr.com/photos/davidascanio/albums/72157706650233041 It’s about 02:00 AM in San José, and we are listening to the widespread and ubiquitous Clay-colored Robin singing outside our hotel windows. Yet, it was still too early to experience the real explosion of bird song, which usually happens after dawn. Then, after 05:30 AM, the chorus started when a vocal Great Kiskadee broke the morning silence, followed by the scratchy notes of two Hoffmann´s Woodpeckers, a nesting pair of Inca Doves, the ascending and monotonous song of the Yellow-bellied Elaenia, and the cacophony of an (apparently!) engaged pair of Rufous-naped Wrens. This was indeed a warm welcome to magical Costa Rica! To complement the first morning of birding, two boreal migrants, Baltimore Orioles and a Tennessee Warbler, joined the bird feast just outside the hotel area. Broad-billed Motmot . Photo: D. Ascanio © Victor Emanuel Nature Tours 2 The Best of Costa Rica, 2019 After breakfast, we drove towards the volcanic ring of Costa Rica. Circling the slope of Poas volcano, we eventually reached the inspiring Bosque de Paz. With its hummingbird feeders and trails transecting a beautiful moss-covered forest, this lodge offered us the opportunity to see one of Costa Rica´s most difficult-to-see Grallaridae, the Scaled Antpitta. -

"Riverside Wren Pairs Jointly Defend Their Territories Against Simulated Intruders"
1 "Riverside wren pairs jointly defend their territories against simulated intruders" 2 (ETH-17-0081.R1) is now accepted for publication in Ethology.” 3 23rd August 2017 4 5 6 7 8 Riverside wren pairs jointly defend their territories against simulated 9 intruders 10 11 Esmeralda Quirós-Guerrero1,4, Maria João Janeiro1,2, Marvin Lopez Morales, Will 12 Cresswell1, Christopher N. Templeton1,3,4 13 14 1 School of Biology, University of St Andrews, St Andrews, United Kingdom. 15 2 CESAM, Department of Biology, University of Aveiro, Aveiro, Portugal. 16 3 Pacific University, Forest Grove, Oregon, United States of America. 17 4Joint corresponding authors; EQ-G email: [email protected]; telephone: (+44) 1334 18 463401; postal address: School of Biology, Sir Harlod Mitchell and Dyres Brae, Greenside 19 Place, St Andrews Fife KY16 9TH, United Kingdom. CNT email: 20 [email protected]; telephone: (+1) 50335 23149; postal address: Pacific University, 21 2043 College Way, Forest Grove, Oregon 97116, United States of America. 22 23 24 25 1 26 Abstract 27 Duets are a jointly-produced signal where two or more individuals coordinate their 28 vocalizations by overlapping or alternating their songs. Duets are used in a wide array of 29 contexts within partnerships, ranging from territory defence to pair bond maintenance. It has 30 been proposed that pairs that coordinate their songs might also better coordinate other 31 activities, including nest building, parental care, and defending shared resources. Here, we 32 test in the riverside wren (Cantorchilus semibadius), a neotropical duetting species that 33 produces highly coordinated duet songs, whether males and females show similar 34 responses to playback. -

Troglodytidae Species Tree
Troglodytidae I Rock Wren, Salpinctes obsoletus Canyon Wren, Catherpes mexicanus Sumichrast’s Wren, Hylorchilus sumichrasti Nava’s Wren, Hylorchilus navai Salpinctinae Nightingale Wren / Northern Nightingale-Wren, Microcerculus philomela Scaly-breasted Wren / Southern Nightingale-Wren, Microcerculus marginatus Flutist Wren, Microcerculus ustulatus Wing-banded Wren, Microcerculus bambla ?Gray-mantled Wren, Odontorchilus branickii Odontorchilinae Tooth-billed Wren, Odontorchilus cinereus Bewick’s Wren, Thryomanes bewickii Carolina Wren, Thryothorus ludovicianus Thrush-like Wren, Campylorhynchus turdinus Stripe-backed Wren, Campylorhynchus nuchalis Band-backed Wren, Campylorhynchus zonatus Gray-barred Wren, Campylorhynchus megalopterus White-headed Wren, Campylorhynchus albobrunneus Fasciated Wren, Campylorhynchus fasciatus Cactus Wren, Campylorhynchus brunneicapillus Yucatan Wren, Campylorhynchus yucatanicus Giant Wren, Campylorhynchus chiapensis Bicolored Wren, Campylorhynchus griseus Boucard’s Wren, Campylorhynchus jocosus Spotted Wren, Campylorhynchus gularis Rufous-backed Wren, Campylorhynchus capistratus Sclater’s Wren, Campylorhynchus humilis Rufous-naped Wren, Campylorhynchus rufinucha Pacific Wren, Nannus pacificus Winter Wren, Nannus hiemalis Eurasian Wren, Nannus troglodytes Zapata Wren, Ferminia cerverai Marsh Wren, Cistothorus palustris Sedge Wren, Cistothorus platensis ?Merida Wren, Cistothorus meridae ?Apolinar’s Wren, Cistothorus apolinari Timberline Wren, Thryorchilus browni Tepui Wren, Troglodytes rufulus Troglo dytinae Ochraceous -

Printable PDF Format
Field Guides Tour Report Safari Brazil: The Pantanal & More 2019 Sep 21, 2019 to Oct 6, 2019 Marcelo Padua & Dan Lane For our tour description, itinerary, past triplists, dates, fees, and more, please VISIT OUR TOUR PAGE. We experienced the amazing termitaria-covered landscape of Emas National Park, where participant Rick Thompson got this evocative image, including two Aplomado Falcons, and a Pampas Deer. Brazil is a big place, and it is home to a wide variety of biomes. Among its most famous are the Amazon and the Pantanal, both occupy huge areas and have their respective hydrologies to thank for their existence. In addition to these are drier regions that cut the humid Amazon from the humid Atlantic Forest, this is known as the “Dry Diagonal,” home to the grasslands we observed at Emas, the chapada de Cipo, and farther afield, the Chaco, Pampas, and Caatinga. We were able to dip our toes into several of these incredible features, beginning with the Pantanal, one of the world’s great wetlands, and home to a wide array of animals, fish, birds, and other organisms. In addition to daytime outings to enjoy the birdlife and see several of the habitats of the region (seasonally flooded grasslands, gallery forest, deciduous woodlands, and open country that does not flood), we were able to see a wide array of mammals during several nocturnal outings, culminating in such wonderful results as seeing multiple big cats (up to three Ocelots and a Jaguar on one night!), foxes, skunks, raccoons, Giant Anteaters, and others. To have such luck as this in the Americas is something special! Our bird list from the region included such memorable events as seeing an active Jabiru nest, arriving at our lodging at Aguape to a crowd of Hyacinth Macaws, as well as enjoying watching the antics of their cousins the Blue-and-yellow Macaws. -

2020 Costa Rica Tour Species List
Costa Rica Guides: Ernesto Carman Eagle-Eye Tours February 22 - March 09, 2020 and Paz Angulo Irola Bird Species - Costa Rica Seen/ Common Name Scientific Name Heard TINAMOUS 1 Great Tinamou Tinamus major 1 2 Little Tinamou Crypturellus soui H DUCKS, GEESE, AND WATERFOWL 3 Black-bellied Whistling-Duck Dendrocygna autumnalis 1 4 Muscovy Duck Cairina moschata 1 5 Blue-winged Teal Anas discors 1 GUANS, CHACHALACAS, AND CURASSOWS 6 Gray-headed Chachalaca Ortalis cinereiceps 1 7 Crested Guan Penelope purpurascens 1 8 Black Guan Chamaepetes unicolor 1 9 Great Curassow Crax rubra 1 NEW WORLD QUAIL 10 Crested Bobwhite Colinus cristatus H 11 Buffy-crowned Wood-Partridge Dendrortyx leucophrys H 12 Spotted Wood-Quail Odontophorus guttatus 1 STORKS 13 Jabiru Jabiru mycteria 1 14 Wood Stork Mycteria americana 1 FRIGATEBIRDS 15 Magnificent Frigatebird Fregata magnificens 1 CORMORANTS AND SHAGS 16 Neotropic Cormorant Phalacrocorax brasilianus 1 ANHINGAS 17 Anhinga Anhinga anhinga 1 PELICANS 18 Brown Pelican Pelecanus occidentalis 1 HERONS, EGRETS, AND BITTERNS 19 Fasciated Tiger-Heron Tigrisoma fasciatum 1 20 Bare-throated Tiger-Heron Tigrisoma mexicanum 1 21 Pinnated Bittern Botaurus pinnatus 1 22 Great Blue Heron Ardea herodias 1 23 Great Egret Ardea alba 1 24 Snowy Egret Egretta thula 1 25 Little Blue Heron Egretta caerulea 1 26 Tricolored Heron Egretta tricolor 1 27 Cattle Egret Bubulcus ibis 1 28 Green Heron Butorides virescens 1 29 Boat-billed Heron Cochlearius cochlearius 1 30 Yellow-crowned Night-Heron Nyctanassa violacea 1 IBISES AND SPOONBILLS -

Panama's Canopy Tower and El Valle's Canopy Lodge
FIELD REPORT – Panama’s Canopy Tower and El Valle’s Canopy Lodge January 4-16, 2019 Orange-bellied Trogon © Ruthie Stearns Blue Cotinga © Dave Taliaferro Geoffroy’s Tamarin © Don Pendleton Ocellated Antbird © Carlos Bethancourt White-tipped Sicklebill © Jeri Langham Prepared by Jeri M. Langham VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DR., AUSTIN, TX 78746 Phone: 512-328-5221 or 800-328-8368 / Fax: 512-328-2919 [email protected] / www.ventbird.com Myriads of magazine articles have touted Panama’s incredible Canopy Tower, a former U.S. military radar tower transformed by Raúl Arias de Para when the U.S. relinquished control of the Panama Canal Zone. It sits atop 900-foot Semaphore Hill overlooking Soberania National Park. While its rooms are rather spartan, the food is Panama’s Canopy Tower © Ruthie Stearns excellent and the opportunity to view birds at dawn from the 360º rooftop Observation Deck above the treetops is outstanding. Twenty minutes away is the start of the famous Pipeline Road, possibly one of the best birding roads in Central and South America. From our base, daily birding outings are made to various locations in Central Panama, which vary from the primary forest around the tower, to huge mudflats near Panama City and, finally, to cool Cerro Azul and Cerro Jefe forest. An enticing example of what awaits visitors to this marvelous birding paradise can be found in excerpts taken from the Journal I write during every tour and later e- mail to participants. These are taken from my 17-page, January 2019 Journal. On our first day at Canopy Tower, with 5 of the 8 participants having arrived, we were touring the Observation Deck on top of Canopy Tower when Ruthie looked up and called my attention to a bird flying in our direction...it was a Black Hawk-Eagle! I called down to others on the floor below and we watched it disappear into the distant clouds. -

Assessing Bird Migrations Verônica Fernandes Gama
Assessing Bird Migrations Verônica Fernandes Gama Master of Philosophy, Remote Sensing Bachelor of Biological Sciences (Honours) A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2019 School of Biological Sciences Abstract Birds perform many types of migratory movements that vary remarkably both geographically and between taxa. Nevertheless, nomenclature and definitions of avian migrations are often not used consistently in the published literature, and the amount of information available varies widely between taxa. Although comprehensive global lists of migrants exist, these data oversimplify the breadth of types of avian movements, as species are classified into just a few broad classes of movements. A key knowledge gap exists in the literature concerning irregular, small-magnitude migrations, such as irruptive and nomadic, which have been little-studied compared with regular, long-distance, to-and- fro migrations. The inconsistency in the literature, oversimplification of migration categories in lists of migrants, and underestimation of the scope of avian migration types may hamper the use of available information on avian migrations in conservation decisions, extinction risk assessments and scientific research. In order to make sound conservation decisions, understanding species migratory movements is key, because migrants demand coordinated management strategies where protection must be achieved over a network of sites. In extinction risk assessments, the threatened status of migrants and non-migrants is assessed differently in the International Union for Conservation of Nature Red List, and the threatened status of migrants could be underestimated if information regarding their movements is inadequate. In scientific research, statistical techniques used to summarise relationships between species traits and other variables are data sensitive, and thus require accurate and precise data on species migratory movements to produce more reliable results. -
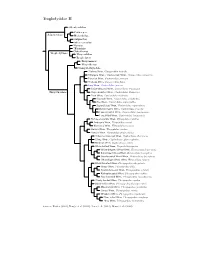
Troglodytidae Tree, Part 2
Troglodytidae II Odontorchilus Catherpes Salpinctinae Hylorchilus Salpinctes Microcerculus Nannus ?Ferminia Cistothorus Troglo dytinae Thryorchilus Troglo dytes Thryomanes Thryothorus Campylorhynchus ?Rufous Wren, Cinnycerthia unirufa ?Sharpe’s Wren / Sepia-brown Wren, Cinnycerthia olivascens Peruvian Wren, Cinnycerthia peruana ?Fulvous Wren, Cinnycerthia fulva Gray Wren, Cantorchilus griseus Stripe-throated Wren, Cantorchilus leucopogon Thryothorinae Stripe-breasted Wren, Cantorchilus thoracicus Plain Wren, Cantorchilus modestus Riverside Wren, Cantorchilus semibadius Bay Wren, Cantorchilus nigricapillus Superciliated Wren, Cantorchilus superciliaris Buff-breasted Wren, Cantorchilus leucotis Fawn-breasted Wren, Cantorchilus guarayanus Long-billed Wren, Cantorchilus longirostris Rufous-and-white Wren, Thryophilus rufalbus Antioquia Wren, Thryophilus sernai Niceforo’s Wren, Thryophilus nicefori Sinaloa Wren, Thryophilus sinaloa Banded Wren, Thryophilus pleurostictus ?Chestnut-breasted Wren, Cyphorhinus thoracicus ?Song Wren, Cyphorhinus phaeocephalus Musician Wren, Cyphorhinus arada White-bellied Wren, Uropsila leucogastra White-breasted Wood-Wren, Henicorhina leucosticta Bar-winged Wood-Wren, Henicorhina leucoptera Gray-breasted Wood-Wren, Henicorhina leucophrys ?Munchique Wood-Wren, Henicorhina negreti Black-throated Wren, Pheugopedius atrogularis Happy Wren, Pheugopedius felix Speckle-breasted Wren, Pheugopedius sclateri Rufous-breasted Wren, Pheugopedius rutilus Spot-breasted Wren, Pheugopedius maculipectus ?Sooty-headed Wren, Pheugopedius spadix Black-bellied Wren, Pheugopedius fasciatoventris Moustached Wren, Pheugopedius genibarbis Coraya Wren, Pheugopedius coraya Whiskered Wren, Pheugopedius mystacalis Plain-tailed Wren, Pheugopedius euophrys ?Inca Wren, Pheugopedius eisenmanni Sources: Barker (2004), Dingle et al. (2006), Lara et al. (2012), Mann et al. (2006).. -

Costa Rica - Bird List
Costa Rica - Bird List Melanerpes formicivorus Acorn Woodpecker Chloroceryle amazona Amazon Kingfisher Coragyps atratus American Black Vulture Fulica americana American Coot Setophaga ruticilla American Redstart Setophaga petechia American Yellow Warbler Anhinga anhinga Anhinga Aratinga astec Aztec Parakeet Icterus galbula Baltimore Oriole Coereba flaveola Bananaquit Campylorhynchus zonatus Band-backed Wren Threnetes ruckeri Band-tailed Barbthroat Tigrisoma mexicanum Bare-throated Tiger-Heron Hirundo rustica Barn Swallow Thamnophilus doliatus Barred Antshrike Falco rufigularis Bat Falcon Cantorchilus nigricapillus Bay Wren Tangara gyrola Bay-headed Tanager Sayornis nigricans Black Phoebe Sporophila corvina Black Seedeater Mniotilta varia Black-and-white Warbler Dendrocygna autumnalis Black-bellied Whistling-Duck Setophaga fusca Blackburnian Warbler Empidonax atriceps Black-capped Flycatcher Basileuterus melanogenys Black-cheeked Warbler Melanerpes pucherani Black-cheeked Woodpecker Icterus prosthemelas Black-cowled Oriole Lophornis helenae Black-crested Coquette Nycticorax nycticorax Black-crowned Night-Heron Tityra inquisitor Black-crowned Tityra Caryothraustes poliogaster Black-faced Grosbeak Myadestes melanops Black-faced Solitaire Catharus mexicanus Black-headed Nightingale-Thrush Saltator atriceps Black-headed Saltator Todirostrum nigriceps Black-headed Tody-Flycatcher Trogon melanocephalus Black-headed Trogon Thamnophilus bridgesi Black-hooded Antshrike Himantopus mexicanus Black-necked Stilt Arremonops conirostris Black-striped -

Short Costa Rica February 22–March 1, 2020
SHORT COSTA RICA FEBRUARY 22–MARCH 1, 2020 Male Resplendent Quetzal © Brian Gibbons LEADERS: BRIAN GIBBONS & MAURICIO CHINCHILLA LIST COMPILED BY: BRIAN GIBBONS VICTOR EMANUEL NATURE TOURS 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM From steamy afternoons in the lowlands to chilly mountain mornings, Costa Rica packs birds, scenery, and tourists into a tiny country. We saw 260 species on our short trip to a couple of the prime birding areas. Netting a few birds on the hotel grounds in the Central Valley, a great haul at La Paz Waterfall Gardens, superb sightings at La Selva, and the crown jewel, Resplendent Quetzal, in the mountains on our last full birding day, Costa Rica produced a wonderful array of avifauna for us. The unchallenged highlight was Terri’s Bare-shanked Screech-Owl that she spotted while we were rattling down a rocky road in an ancient Land Cruiser among the moss-encrusted oaks of the cloudforest! A Pair of Prong-billed Barbets at La Paz Waterfall Garden © Brian Gibbons Starting in the Central Valley on a windy morning, we tallied a few birds that we wouldn’t see during the rest of the tour. Hoffman’s Woodpecker, Rufous-naped Wren, Cabanis’s Wren, and the Blue-vented Hummingbird (formerly Steely-vented) all made an appearance in the wind. Tennessee Warblers were also very much in evidence around the fig tree that attracted so much attention, readying for the trip north. The feeders at La Paz threatened to take all of our time from the gardens; hummingbirds and tanagers were abundant—often at arm’s-length. -

Henicorhina Leucosticta)
UNIVERSITY OF CALIFORNIA Los Angeles Complex singing behavior of the White-breasted Wood Wren (Henicorhina leucosticta) A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biology by Zachary Thomas Harlow 2013 © Copyright by Zachary Thomas Harlow 2013 ABSTRACT OF THE DISSERTATION Complex singing behavior of the White-breasted Wood Wren (Henicorhina leucosticta) by Zachary Thomas Harlow Doctor of Philosophy in Biology University of California, Los Angeles, 2013 Professor Charles E. Taylor, Chair Neotropical bird species provide exceptional opportunities for advancing the understanding of avian communication systems. This dissertation focuses on the singing behavior of the White-breasted Wood Wren (Henicorhina leucosticta), which is not well understood even though it expresses behaviors such as female song and duetting that are at the forefront of research in avian communication. I present the background in this area of research and make the case that this species is of special interest to the study of avian communication because of its phylogenetic relationship to other duetting wrens and because its singing style is not easily categorized by singing behavior of other duetting species. In Chapter 2, I characterize the repertoire and singing behavior of this species ii and show that male repertoires are larger than those of females and can be classified into three groups based on spectral characteristics and song use. I describe unique high- frequency ‘introductory notes’ that directly precede songs within a bout but are facultatively included with the songs. In Chapter 3, I discuss playback experiments that suggest introductory notes act as a graded signal in certain song types during countersinging interactions. -

A Duetting Perspective on Avian Song Learning ⁎ Karla D
Behavioural Processes xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Behavioural Processes journal homepage: www.elsevier.com/locate/behavproc A duetting perspective on avian song learning ⁎ Karla D. Rivera-Cáceresa, Christopher N. Templetronb,c, a Department of Biology, University of Miami, Coral Gables, FL, USA b Department of Biology, Pacific University, Forest Grove, OR, USA c School of Biology, University of St Andrews, St Andrews, Scotland, UK ARTICLE INFO ABSTRACT Keywords: Avian song learning has a rich history of study and has become the preeminent system for understanding the Auditory-forebrain pathway ontogeny of vocal communication in animals. Song learning in birds has many parallels with human language Avian duets learning, ranging from the neural mechanisms involved to the importance of social factors in shaping signal Conversation acquisition. While much has been learned about the process of song learning, virtually all of the research done to Duetting date has focused on temperate species, where often only one sex (the male) sings. Duetting species, in which both Duet learning males and females learn to sing and learn to combine their songs into temporally coordinated joint displays, Language Song learning could provide many insights into the processes by which vocal learning takes place. Here we highlight three key Vocal interactions features of song learning—neuroendocrine control mechanisms, timing and life history stages of song acquisi- tion, and the role of social factors in song selection and use—that have been elucidated from species where only males sing, and compare these with duetting species. We summarize what is known about song learning in duetting species and then provide several suggestions for fruitful directions for future research.