Protein Expression Pattern in Response to Ionizing Radiation in MCF-7 Human Breast Cancer Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic and Pharmacological Approaches to Preventing Neurodegeneration
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Genetic and Pharmacological Approaches to Preventing Neurodegeneration Marco Boccitto University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Neuroscience and Neurobiology Commons Recommended Citation Boccitto, Marco, "Genetic and Pharmacological Approaches to Preventing Neurodegeneration" (2012). Publicly Accessible Penn Dissertations. 494. https://repository.upenn.edu/edissertations/494 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/494 For more information, please contact [email protected]. Genetic and Pharmacological Approaches to Preventing Neurodegeneration Abstract The Insulin/Insulin-like Growth Factor 1 Signaling (IIS) pathway was first identified as a major modifier of aging in C.elegans. It has since become clear that the ability of this pathway to modify aging is phylogenetically conserved. Aging is a major risk factor for a variety of neurodegenerative diseases including the motor neuron disease, Amyotrophic Lateral Sclerosis (ALS). This raises the possibility that the IIS pathway might have therapeutic potential to modify the disease progression of ALS. In a C. elegans model of ALS we found that decreased IIS had a beneficial effect on ALS pathology in this model. This beneficial effect was dependent on activation of the transcription factor daf-16. To further validate IIS as a potential therapeutic target for treatment of ALS, manipulations of IIS in mammalian cells were investigated for neuroprotective activity. Genetic manipulations that increase the activity of the mammalian ortholog of daf-16, FOXO3, were found to be neuroprotective in a series of in vitro models of ALS toxicity. -

Apoptotic Genes As Potential Markers of Metastatic Phenotype in Human Osteosarcoma Cell Lines
17-31 10/12/07 14:53 Page 17 INTERNATIONAL JOURNAL OF ONCOLOGY 32: 17-31, 2008 17 Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines CINZIA ZUCCHINI1, ANNA ROCCHI2, MARIA CRISTINA MANARA2, PAOLA DE SANCTIS1, CRISTINA CAPANNI3, MICHELE BIANCHINI1, PAOLO CARINCI1, KATIA SCOTLANDI2 and LUISA VALVASSORI1 1Dipartimento di Istologia, Embriologia e Biologia Applicata, Università di Bologna, Via Belmeloro 8, 40126 Bologna; 2Laboratorio di Ricerca Oncologica, Istituti Ortopedici Rizzoli; 3IGM-CNR, Unit of Bologna, c/o Istituti Ortopedici Rizzoli, Via di Barbiano 1/10, 40136 Bologna, Italy Received May 29, 2007; Accepted July 19, 2007 Abstract. Metastasis is the most frequent cause of death among malignant primitive bone tumor, usually developing in children patients with osteosarcoma. We have previously demonstrated and adolescents, with a high tendency to metastasize (2). in independent experiments that the forced expression of Metastases in osteosarcoma patients spread through peripheral L/B/K ALP and CD99 in U-2 OS osteosarcoma cell lines blood very early and colonize primarily the lung, and later markedly reduces the metastatic ability of these cancer cells. other skeleton districts (3). Since disseminated hidden micro- This behavior makes these cell lines a useful model to assess metastases are present in 80-90% of OS patients at the time the intersection of multiple and independent gene expression of diagnosis, the identification of markers of invasiveness signatures concerning the biological problem of dissemination. and metastasis forms a target of paramount importance in With the aim to characterize a common transcriptional profile planning the treatment of osteosarcoma lesions and enhancing reflecting the essential features of metastatic behavior, we the prognosis. -

Role and Regulation of Snon/Skil and PLSCR1 Located at 3Q26.2
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School 9-18-2014 Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology Madhav Karthik Kodigepalli University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Cell Biology Commons, Microbiology Commons, and the Molecular Biology Commons Scholar Commons Citation Kodigepalli, Madhav Karthik, "Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology" (2014). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/5426 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology by Madhav Karthik Kodigepalli A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Cell and Molecular Biology Department of Cell Biology, Microbiology and Molecular Biology College of Arts and Sciences University of South Florida Major Professor: Meera Nanjundan, Ph.D. Richard Pollenz, Ph.D. Patrick Bradshaw, Ph.D. Sandy Westerheide, Ph.D. Date of Approval: September 18, 2014 Keywords: Chemotherapeutics, phospholipid scramblase, toll-like receptor, interferon, dsDNA Copyright © 2014, Madhav Karthik Kodigepalli Dedication I dedicate this research at the lotus feet of Bhagwan Sri Sathya Sai Baba and all the Masters for I am what I am due to their divine grace. -

Analysis of Sequence Architecture at Human-Gibbon Synteny Breaks
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Girirajan et al Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites Santhosh Girirajan1*, Lin Chen1*, Tina Graves2, Tomas Marques-Bonet1, Mario Ventura3, Catrina Fronick2, Lucinda Fulton2, Mariano Rocchi3, Robert S. Fulton2, Richard K. Wilson2, Elaine R. Mardis2 and Evan E. Eichler1† 1. Department of Genome Sciences, Howard Hughes Medical Institute, University of Washington School of Medicine, Seattle, Washington, USA 2. Genome Sequencing Center, Washington University, St. Louis, Missouri, USA 3. Department of Genetics and Microbiology, University of Bari, 70126 Bari, Italy *These authors contributed equally to work †Corresponding author: Evan E. Eichler, PhD University of Washington School of Medicine Howard Hughes Medical Institute Box 355065 Foege S413C, 1705 NE Pacific St. Seattle, WA 98195 E-mail: [email protected] 1 Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Girirajan et al ABSTRACT The gibbon genome exhibits extensive karyotypic diversity with an increased rate of chromosomal rearrangements during evolution. In an effort to understand the mechanistic origin and implications of these rearrangement events, we sequenced 24 synteny breakpoint regions in the white-cheeked gibbon (Nomascus leucogenys, NLE) in the form of high-quality BAC insert sequences (4.2 Mbp). While there is a significant deficit of breakpoints in genes, we identified seven human gene structures, involved in signaling pathways (DEPDC4, GNG10), phospholipid metabolism (ENPP5, PLSCR2), β-oxidation (ECH1), cellular structure and transport (HEATR4), and transcription (GIOT1), that have been disrupted in the NLE gibbon lineage. -

Table S2.A-B. Differentially Expressed Genes Following Activation of OGR1 by Acidic Ph in Mouse Peritoneal Macrophages Ph 6.7 24 H
Table S2.A-B. Differentially expressed genes following activation of OGR1 by acidic pH in mouse peritoneal macrophages pH 6.7 24 h. A. Gene List, including gene process B. Complete Table (Excel). Rank Symbol Full name Involved in: WT/KO (Reference: Gene Card, NCBI, JAX, Uniprot, Ratio unless otherwise indicated) 1. Cyp11a1 Cholesterol side chain cleavage Cholesterol, lipid or steroid metabolism. enzyme, mitochondrial (Cytochrome Catalyses the side-chain cleavage reaction of P450 11A1) cholesterol to pregnenolone. 2. Sparc Secreted acidic cysteine rich Cell adhesion, wound healing, ECM glycoprotein (Osteonectin, Basement interactions, bone mineralization. Activates membrane protein 40 (BM-40)) production and activity of matrix metalloproteinases. 3. Tpsb2 Tryptase beta-2 or tryptase II (trypsin- Inflammatory response, proteolysis. like serine protease) 4. Inhba Inhibin Beta A or Activin beta-A chain Immune response and mediators of inflammation and tissue repair.2-5 5. Cpe Carboxypeptidase E Insulin processing, proteolysis. 6. Igfbp7 Insulin-like growth factor-binding Stimulates prostacyclin (PGI2) production and protein 7 cell adhesion. Induced by retinoic acid. 7. Clu Clusterin Chaperone-mediated protein folding, positive regulation of NF-κB transcription factor activity. Protects cells against apoptosis and cytolysis by complement. Promotes proteasomal degradation of COMMD1 and IKBKB. 8. Cma1 Chymase 1 Cellular response to glucose stimulus, interleukin-1 beta biosynthetic process. Possible roles: vasoactive peptide generation, extracellular matrix degradation. 9. Sfrp4 Secreted frizzled-related protein 4 Negative regulation of Wnt signalling. Increases apoptosis during ovulation. Phosphaturic effects by specifically inhibiting sodium-dependent phosphate uptake. 10. Ephx2 Bifunctional epoxide hydrolase Cholesterol homeostasis, xenobiotic metabolism by degrading potentially toxic epoxides. -

Mouse Hnrnph3 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Hnrnph3 Knockout Project (CRISPR/Cas9) Objective: To create a Hnrnph3 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Hnrnph3 gene (NCBI Reference Sequence: NM_001079824 ; Ensembl: ENSMUSG00000020069 ) is located on Mouse chromosome 10. 10 exons are identified, with the ATG start codon in exon 2 and the TGA stop codon in exon 10 (Transcript: ENSMUST00000020263). Exon 2~3 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 2 starts from the coding region. Exon 2~3 covers 24.18% of the coding region. The size of effective KO region: ~768 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 10 Legends Exon of mouse Hnrnph3 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. Tandem repeats are found in the dot plot matrix. The gRNA site is selected outside of these tandem repeats. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 534 bp section downstream of Exon 3 is aligned with itself to determine if there are tandem repeats. -

Supplementary Tables S1-S3
Supplementary Table S1: Real time RT-PCR primers COX-2 Forward 5’- CCACTTCAAGGGAGTCTGGA -3’ Reverse 5’- AAGGGCCCTGGTGTAGTAGG -3’ Wnt5a Forward 5’- TGAATAACCCTGTTCAGATGTCA -3’ Reverse 5’- TGTACTGCATGTGGTCCTGA -3’ Spp1 Forward 5'- GACCCATCTCAGAAGCAGAA -3' Reverse 5'- TTCGTCAGATTCATCCGAGT -3' CUGBP2 Forward 5’- ATGCAACAGCTCAACACTGC -3’ Reverse 5’- CAGCGTTGCCAGATTCTGTA -3’ Supplementary Table S2: Genes synergistically regulated by oncogenic Ras and TGF-β AU-rich probe_id Gene Name Gene Symbol element Fold change RasV12 + TGF-β RasV12 TGF-β 1368519_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 42.22 5.53 75.28 1373000_at sushi-repeat-containing protein, X-linked 2 (predicted) Srpx2 19.24 25.59 73.63 1383486_at Transcribed locus --- ARE 5.93 27.94 52.85 1367581_a_at secreted phosphoprotein 1 Spp1 2.46 19.28 49.76 1368359_a_at VGF nerve growth factor inducible Vgf 3.11 4.61 48.10 1392618_at Transcribed locus --- ARE 3.48 24.30 45.76 1398302_at prolactin-like protein F Prlpf ARE 1.39 3.29 45.23 1392264_s_at serine (or cysteine) peptidase inhibitor, clade E, member 1 Serpine1 ARE 24.92 3.67 40.09 1391022_at laminin, beta 3 Lamb3 2.13 3.31 38.15 1384605_at Transcribed locus --- 2.94 14.57 37.91 1367973_at chemokine (C-C motif) ligand 2 Ccl2 ARE 5.47 17.28 37.90 1369249_at progressive ankylosis homolog (mouse) Ank ARE 3.12 8.33 33.58 1398479_at ryanodine receptor 3 Ryr3 ARE 1.42 9.28 29.65 1371194_at tumor necrosis factor alpha induced protein 6 Tnfaip6 ARE 2.95 7.90 29.24 1386344_at Progressive ankylosis homolog (mouse) -

Endocrine System Local Gene Expression
Copyright 2008 By Nathan G. Salomonis ii Acknowledgments Publication Reprints The text in chapter 2 of this dissertation contains a reprint of materials as it appears in: Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007 Jun 24;8:218. The co-authors listed in this publication co-wrote the manuscript (AP and KH) and provided critical feedback (see detailed contributions at the end of chapter 2). The text in chapter 3 of this dissertation contains a reprint of materials as it appears in: Salomonis N, Cotte N, Zambon AC, Pollard KS, Vranizan K, Doniger SW, Dolganov G, Conklin BR. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6(2):R12. Epub 2005 Jan 28. The co-authors listed in this publication developed the hierarchical clustering method (KP), co-designed the study (NC, AZ, BC), provided statistical guidance (KV), co- contributed to GenMAPP 2.0 (SD) and performed quantitative mRNA analyses (GD). The text of this dissertation contains a reproduction of a figure from: Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. Epub 2004 Sep 13. The reproduction was taken without permission (chapter 1), figure 1.3. iii Personal Acknowledgments The achievements of this doctoral degree are to a large degree possible due to the contribution, feedback and support of many individuals. To all of you that helped, I am extremely grateful for your support. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -
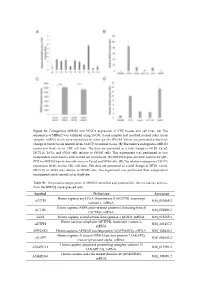
Figure S1. Endogenous MIR45
Figure S1. Endogenous MIR452 and VEGFA expression in CRC tissues and cell lines. (A) The expression of MIR452 was validated using 10 CRC tissue samples and matched normal colon tissue samples. miRNA levels were normalized to colon-specific RNU48. Values are presented as the fold- change in tumor tissue relative levels (ΔΔCT) to normal tissue. (B) The relative endogenous MIR452 expression levels in six CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, LoVo, and SW48 cells relative to SW480 cells. This experiment was performed as two independent experiments, each carried out in triplicate. (C) MIR452 expression level analysis by qRT- PCR for MIR452 transfection efficiency in Caco2 and SW48 cells. (D) The relative endogenous VEGFA expression levels in five CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, or SW48 cells relative to SW480 cells. This experiment was performed three independent experiments, each carried out in duplicate. Table S1. The putative target genes of MIR452 identified and predicted by the microarray analysis from the MIR452 overexpressed cells. Symbol Definition Accession Homo sapiens acyl-CoA thioesterase 8 (ACOT8), transcript ACOT8 NM_005469.2 variant 1, mRNA. Homo sapiens ARP6 actin-related protein 6 homolog (yeast) ACTR6 NM_022496.3 (ACTR6), mRNA. ADI1 Homo sapiens acireductone dioxygenase 1 (ADI1), mRNA. NM_018269.1 Homo sapiens aftiphilin (AFTPH), transcript variant 1, AFTPH NM_203437.2 mRNA. AHNAK2 Homo sapiens AHNAK nucleoprotein 2 (AHNAK2), mRNA. NM_138420.2 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), AKAP7 NM_004842.2 transcript variant alpha, mRNA. Homo sapiens anaphase promoting complex subunit 13 ANAPC13 NM_015391.2 (ANAPC13), mRNA. -

Early Pregnancy-Induced Transcripts in Peripheral Blood Immune Cells In
www.nature.com/scientificreports OPEN Early pregnancy‑induced transcripts in peripheral blood immune cells in Bos indicus heifers Cecilia Constantino Rocha1, Sónia Cristina da Silva Andrade2, Gabriela Dalmaso de Melo1, Igor Garcia Motta1, Luiz Lehmann Coutinho3, Angela Maria Gonella‑Diaza4, Mario Binelli5 & Guilherme Pugliesi1* Immune cells play a central role in early pregnancy establishment in cattle. We aimed to: (1) discover novel early-pregnancy-induced genes in peripheral blood mononuclear cells (PBMC); and (2) characterize the temporal pattern of early‑pregnancy‑induced transcription of select genes in PBMC and peripheral blood polymorphonuclear cells (PMN). Beef heifers were artifcially inseminated on D0 and pregnancies were diagnosed on D28. On D10, 14, 16, 18, and 20, blood was collected for isolation of PBMC and PMN from heifers that were retrospectively classifed as pregnant (P) or non-pregnant (NP). PBMC samples from D18 were submitted to RNAseq and 220 genes were diferentially expressed between pregnant (P) and non-pregnant (NP) heifers. The temporal abundance of 20 transcripts was compared between P and NP, both in PBMC and PMN. In PBMC, pregnancy stimulated transcription of IFI6, RSAD2, IFI44, IFITM2, CLEC3B, OAS2, TNFSF13B, DMKN and LGALS3BP as early as D18. Expression of IFI44, RSAD2, OAS2, LGALS3BP, IFI6 and C1R in PMN was stimulated in the P group from D18. The novel early-pregnancy induced genes discovered in beef heifers will allow both the understanding of the role of immune cells during the pre‑attachment period and the development of technologies to detect early pregnancies in beef cattle. In cattle, pregnancy success depends on the maintenance of a functional corpus luteum (CL) beyond the time of luteolysis, which normally occurs between days 15 and 18 of the estrous cycle 1,2. -

(12) Patent Application Publication (10) Pub. No.: US 2003/0170678A1 Tanzi Et Al
US 2003.017O678A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0170678A1 Tanzi et al. (43) Pub. Date: Sep. 11, 2003 (54) GENETIC MARKERS FOR ALZHEIMER'S Related U.S. Application Data DISEASE AND METHODS USING THE SAME (60) Provisional application No. 60/348,065, filed on Oct. (75) Inventors: Rudolph E. Tanzi, Hull, MA (US); 25, 2001. Provisional application No. 60/336,983, Kenneth David Becker, San Diego, CA filed on Nov. 2, 2001. (US); Gonul Velicelebi, San Diego, CA (US); Kathryn J. Elliott, San Diego, Publication Classification CA (US); Xin Wang, San Diego, CA (US); Lars Bertram, Brighton, MA (51) Int. C.7 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - C12O 1/68 (US); Aleister J. Saunders, (52) U.S. Cl. .................................................................. 435/6 Philadelphia, PA (US); Deborah Lynne Blacker, Newton, MA (US) (57) ABSTRACT Correspondence Address: HELLER EHRMAN WHITE & MCAULIFFE LLP Genetic markers associated with Alzheimer's disease are 4350 LA JOLLAVILLAGE DRIVE provided. Also provided are methods of determining the 7TH FLOOR presence or absence in a Subject of one or more polymor SAN DIEGO, CA 92122-1246 (US) phisms associated with Alzheimer's disease and methods of determining the level of risk for Alzheimer's disease in a (73) Assignee: Neurogenetics, Inc. Subject. Further provided are nucleic acid compositions and kits for use in determining the presence or absence in a (21) Appl. No.: 10/281,456 Subject of one or more polymorphisms associated with Alzheimer's disease and kits for determining the level of (22) Filed: Oct. 25, 2002 risk for Alzheimer's disease in a Subject. Patent Application Publication Sep.