Early Pregnancy-Induced Transcripts in Peripheral Blood Immune Cells In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Rapid and Effective Nonsurgical Artificial Insemination Protocol Using
Transgenic Res (2015) 24:775–781 DOI 10.1007/s11248-015-9887-3 TECHNICAL REPORT A rapid and effective nonsurgical artificial insemination protocol using the NSETTM device for sperm transfer in mice without anesthesia Barbara J. Stone . Kendra H. Steele . Angelika Fath-Goodin Received: 27 January 2015 / Accepted: 3 June 2015 / Published online: 12 June 2015 Ó The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Artificial insemination (AI) is an assisted Introduction reproductive technique that is implemented success- fully in humans as a fertility treatment, performed Artificial insemination (AI) is used to assist in extensively for commercial breeding of livestock, and reproduction by directly delivering motile sperm to is also successful in laboratory rodents. AI in the the female reproductive tract. Artificial insemination mouse may be especially useful for breeding of in mice can be useful as an alternative to breeding for transgenic or mutant mice with fertility problems, strains which are not easily bred due to physical or expansion of mouse colonies, and as an alternative to behavioral issues. The techniques of AI and in vitro in vitro fertilization. Nonsurgical AI techniques for the fertilization (IVF) can both be used to generate viable mouse have been described previously but are not embryos. Development after AI simply continues in often implemented due to technical difficulties. Here the recipient female, while embryos generated from we compare various protocols for preparation of CD1 IVF must be transferred to a pseudopregnant recipient. recipients prior to AI for naı¨ve (in estrus), ovulation- AI or IVF can be used to generate embryos after induced, and superovulated females. -

The Influence of Progesterone on Bovine Uterine Fluid Energy
www.nature.com/scientificreports OPEN The infuence of progesterone on bovine uterine fuid energy, nucleotide, vitamin, cofactor, Received: 5 February 2019 Accepted: 7 May 2019 peptide, and xenobiotic Published: xx xx xxxx composition during the conceptus elongation-initiation window Constantine A. Simintiras, José M. Sánchez, Michael McDonald & Patrick Lonergan Conceptus elongation coincides with one of the periods of greatest pregnancy loss in cattle and is characterized by rapid trophectoderm expansion, commencing ~ Day 13 of pregnancy, i.e. before maternal pregnancy recognition. The process has yet to be recapitulated in vitro and does not occur in the absence of uterine gland secretions in vivo. Moreover, conceptus elongation rates are positively correlated to systemic progesterone in maternal circulation. It is, therefore, a maternally-driven and progesterone-correlated developmental phenomenon. This study aimed to comprehensively characterize the biochemical composition of the uterine luminal fuid on Days 12–14 – the elongation- initiation window – in heifers with normal vs. high progesterone, to identify molecules potentially involved in conceptus elongation initiation. Specifcally, nucleotide, vitamin, cofactor, xenobiotic, peptide, and energy metabolite profles of uterine luminal fuid were examined. A total of 59 metabolites were identifed, of which 6 and 3 displayed a respective progesterone and day efect, whereas 16 exhibited a day by progesterone interaction, of which 8 were nucleotide metabolites. Corresponding pathway enrichment -

The Artificial Way
SPOTLIGHT Breeding Rhinos for the future The Artificial way BY FELIX PATTON aptive breeding programmes have many decades about the husbandry and had some spectacular successes in reproductive needs of captive Black and 310 Csaving species and reintroducing Indian rhinos, when applied to White them into the wild. All rhino species Number of oestrous cycles rhinos, it has been unsuccessful. The are at risk from poaching, disease and non-reproducing White rhino main problem has been the absence of or natural disaster. Developing captive female in captivity can exhibit. erratic nature of oestrous cycle activity in populations is an essential safeguard to over half of the females in the European avoid extinction but breeding success has and North American Species Survival been limited. If a female rhino does not Program. get pregnant, her uterus starts to develop White rhino females in the wild irreversible problems, such as cysts To date, captive reproduction in usually experience short intervals and tumours. Can the failure to become the White rhino has been poor. Despite between successive births, even as pregnant be overcome? the wealth of knowledge acquired over little as 18 months. This indicates that pregnancy and lactation are the most common endocrine profile with possibly as few as 30 oestrous cycles per reproductive life span. Ultrasound technology for use in rhinoceros has been A reproducing female White rhino in used in well over 150 reproductive assessments in more captivity may produce up to nine calves. With a pregnancy of 16 months and than 70 rhinos with the result that the causes of poor subsequent lactation of approximately captive performance are now known. -

Artificial Insemination
Ch36-A03309.qxd 1/23/07 5:16 PM Page 539 Section 6 Infertility and Recurrent Pregnancy Loss Chapter Artificial Insemination 36 Ashok Agarwal and Shyam S. R. Allamaneni INTRODUCTION widely available, the terms homologous artificial insemination and heterologous artificial insemination were used to differentiate Artificial insemination is an assisted conception method that can these two alternative sources. However, the use of these bio- be used to alleviate infertility in selected couples. The rationale medical terms in this manner is at variance with their scientific behind the use of artificial insemination is to increase the gamete meaning, where they denote different species or organisms (as in, density near the site of fertilization.1 The effectiveness of artificial e.g., homologous and heterologous tissue grafts). insemination has been clearly established in specific subsets of In the latter half of the 20th century, the terms artificial infertile patients such as those with idiopathic infertility, infertility insemination, donor (AID) and artificial insemination, husband related to a cervical factor, or a mild male factor infertility (AIH) found common use. However, the widespread use of the (Table 36-1).2,3 An accepted advantage of artificial insemination acronym AIDS for acquired immunodeficiency syndrome resulted is that it is generally less expensive and invasive than other in the replacement of AID with therapeutic donor insemination assisted reproductive technology (ART) procedures.4 (TDI). An analogous alternative term for AIH has not evolved, This chapter provides a comprehensive description of probably in part because of the increasingly common situation indications for artificial insemination, issues to consider before where the woman’s partner is not her legal husband. -

In Vitro Fertilization and Artificial Insemination: Ethical Consideration
Loyola University Chicago Loyola eCommons Dissertations Theses and Dissertations 1997 In Vitro Fertilization and Artificial Insemination: Ethical Consideration Joseph Ibegbulem Ekweariri Loyola University Chicago Follow this and additional works at: https://ecommons.luc.edu/luc_diss Part of the Philosophy Commons Recommended Citation Ekweariri, Joseph Ibegbulem, "In Vitro Fertilization and Artificial Insemination: Ethical Consideration" (1997). Dissertations. 3716. https://ecommons.luc.edu/luc_diss/3716 This Dissertation is brought to you for free and open access by the Theses and Dissertations at Loyola eCommons. It has been accepted for inclusion in Dissertations by an authorized administrator of Loyola eCommons. For more information, please contact [email protected]. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 License. Copyright © 1997 Joseph Ibegbulem Ekweariri LOYOLA UNIVERSITY OF CHICAGO IN VITRO FERTILIZATION AND ARTIFICIAL INSEMINATION: ETHICAL CONSIDERATION A DISSERTATION SUBMITTED TO THEFACULTYOFTHEGRADUATESCHOOL IN CANDIDACY FOR THE DEGREE OF DOCTOR OF PHILOSOPHY DEPARTMENT OF PHILOSOPHY BY JOSEPH IBEGBULEM EKWEARIRI CHICAGO, ILLINOIS MAY, 1997 Copyright by Joseph Ibegbulem Ekweariri, 1997 All rights reserved. ACKNOWLEDGMENTS I thank you God for my life; even when ill-health threatened my studies and this work you sustained me throughout. Many people contributed to the success of this work in different capacities. I wish to thank them. I am grateful to my dissertation committee: Prof. David T. Ozar, the Chairman of the committee and director of my dissertation, Prof. Richard Westley, and Prof. John Langan S.J. I wish to thank all my professors at Loyola University, especially Prof. Kenneth Thompson, the director of the ~raduate School of Philosophy and Max Caproni Asst. -

The Protection of the Human Embryo in Vitro
Strasbourg, 19 June 2003 CDBI-CO-GT3 (2003) 13 STEERING COMMITTEE ON BIOETHICS (CDBI) THE PROTECTION OF THE HUMAN EMBRYO IN VITRO Report by the Working Party on the Protection of the Human Embryo and Fetus (CDBI-CO-GT3) Table of contents I. General introduction on the context and objectives of the report ............................................... 3 II. General concepts............................................................................................................................... 4 A. Biology of development ....................................................................................................................... 4 B. Philosophical views on the “nature” and status of the embryo............................................................ 4 C. The protection of the embryo............................................................................................................... 8 D. Commercialisation of the embryo and its parts ................................................................................... 9 E. The destiny of the embryo ................................................................................................................... 9 F. “Freedom of procreation” and instrumentalisation of women............................................................10 III. In vitro fertilisation (IVF).................................................................................................................. 12 A. Presentation of the procedure ...........................................................................................................12 -

Actions of Interferon Tau During Maternal Recognition of Pregnancy Ações Do Interferon Tau Durante O Reconhecimento Materno Da Gestação
Anais do XXIII Congresso Brasileiro de Reprodução Animal (CBRA-2019); Gramado, RS, 15 a 17 de maio de 2019. Actions of interferon tau during maternal recognition of pregnancy Ações do interferon tau durante o reconhecimento materno da gestação Alfredo Quites Antoniazzi1, £, Carolina dos Santos Amaral1, Alessandra Brid2 1BioRep – Laboratory of Biotechnology and Animal Reproduction, Federal University of Santa Maria, Santa Maria, RS, Brazil. 2Department of Veterinary Medicine, Faculty of Animal Sciences and Food Engineering, University of São Paulo, Pirassununga, SP, Brazil. Abstract Maternal recognition of pregnancy in ruminants is a physiological process that requires an interaction between the conceptus and the mother in order to avoid luteal regression. Studies on 60’s started to investigate the communication, and late on the 80’s it was identified a Type I interferon called interferon tau. In the 90’s the local action of interferon tau was described. It suppresses luteolytic pulses of prostaglandin F2 alpha inhibiting endometrial expression of estrogen and oxytocin receptors. Several studies presented the expression of interferon- stimulated genes on extrauterine tissues. After, the endocrine action of interferon tau was described. This study revealed a greater interferon bioactivity in the serum of uterine vein from pregnant ewes when compared to non- pregnant ewes. Following a series of experiments were preformed to better understand the endocrine action of interferon tau in several extrauteine tissues. This review presents the endocrine action of interferon tau during maternal recognition of pregnancy period in ruminants. Keywords: IFNT, pregnancy, endocrine, paracrine, interferon-stimulated genes, ruminants. Resumo O reconhecimento materno da gestação em ruminantes é um processo fisiológico que requer a interação entre o concepto e a mãe com o obejetivo de evitar a regressão luteal. -

Role and Regulation of Snon/Skil and PLSCR1 Located at 3Q26.2
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School 9-18-2014 Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology Madhav Karthik Kodigepalli University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Cell Biology Commons, Microbiology Commons, and the Molecular Biology Commons Scholar Commons Citation Kodigepalli, Madhav Karthik, "Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology" (2014). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/5426 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role and Regulation of SnoN/SkiL and PLSCR1 Located at 3q26.2 and 3q23, Respectively, in Ovarian Cancer Pathophysiology by Madhav Karthik Kodigepalli A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Cell and Molecular Biology Department of Cell Biology, Microbiology and Molecular Biology College of Arts and Sciences University of South Florida Major Professor: Meera Nanjundan, Ph.D. Richard Pollenz, Ph.D. Patrick Bradshaw, Ph.D. Sandy Westerheide, Ph.D. Date of Approval: September 18, 2014 Keywords: Chemotherapeutics, phospholipid scramblase, toll-like receptor, interferon, dsDNA Copyright © 2014, Madhav Karthik Kodigepalli Dedication I dedicate this research at the lotus feet of Bhagwan Sri Sathya Sai Baba and all the Masters for I am what I am due to their divine grace. -

Interferon Receptor Signaling in Malignancy: a Network of Cellular Pathways Defining Biological Outcomes
Published OnlineFirst September 12, 2014; DOI: 10.1158/1541-7786.MCR-14-0450 Molecular Cancer Review Research Interferon Receptor Signaling in Malignancy: A Network of Cellular Pathways Defining Biological Outcomes Eleanor N. Fish1 and Leonidas C. Platanias2 Abstract IFNs are cytokines with important antiproliferative activity and exhibit key roles in immune surveillance against malignancies. Early work initiated over three decades ago led to the discovery of IFN receptor activated Jak–Stat pathways and provided important insights into mechanisms for transcriptional activation of IFN- stimulated genes (ISG) that mediate IFN biologic responses. Since then, additional evidence has established critical roles for other receptor-activated signaling pathways in the induction of IFN activities. These include MAPK pathways, mTOR cascades, and PKC pathways. In addition, specific miRNAs appear to play a significant role in the regulation of IFN signaling responses. This review focuses on the emerging evidence for a model in which IFNs share signaling elements and pathways with growth factors and tumorigenic signals but engage them in a distinctive manner to mediate antiproliferative and antiviral responses. Mol Cancer Res; 12(12); 1691–703. Ó2014 AACR. Introduction as aplastic anemia (6). There is also recent evidence for Because of their antineoplastic, antiviral, and immuno- opposing actions of distinct IFN subtypes in the patho- modulatory properties, recombinant IFNs have been used physiology of certain diseases. For instance, a recent study extensively in the treatment of various diseases in humans demonstrated that there is an inverse association between (1). IFNs have clinical activity against several malignancies IFNb and IFNg gene expression in human leprosy, con- and are actively used in the treatment of solid tumors such sistent with opposing functions between type I and II as malignant melanoma and renal cell carcinoma; and IFNs in the pathophysiology of this disease (7). -

Assisted Reproduction in Developing Countries—Facing up to the Issues
1 Progress, No. 63 No. 63 2003 UNDP/UNFPA/WHO/World Assisted reproduction in developing Bank Special Programme of Research, Development and countriesfacing up to the issues Research Training in Human Reproduction (HRP) For millions of couples around the Attitudes to assisted reproduction are Department of Reproductive world, the inability to have children is very much influenced by the specific Health and Research, a personal tragedy. For a significant cultural and social context. Manipula- World Health Organization, proportion of them, the private agony tion of sperm, oocytes and embryos Geneva, Switzerland is compounded by a social stigma, raises numerous questions to which which can have serious and far-reach- there are no easy answers, and which Launched by the World Health ing consequences. It is not surprising often arouse strong emotions. The Organization in 1972, the UNDP/ therefore that the demand for as- article on page 5 outlines some of the UNFPA/WHO/World Bank sisted reproductive technologies ethical and legal issues surrounding Special Programme of (ART) is growing in all regions. But the use of ART, which need to be Research, Development and how can the provision of these tech- discussed openly by all involved par- Research Training in Human nologieswhich are expensive and ties wherever such services are made Reproduction is a global have a success rate of less than available. programme of technical 30%be justified in developing coun- cooperation. It promotes, tries with poorly developed health In the 25 years since the birth of the coordinates, supports, conducts, services that are still struggling with first baby resulting from in vitro ferti- and evaluates research on infectious and chronic diseases, such lization, there have been tremendous reproductive health, with as tuberculosis, malaria and HIV/ advances in scientific techniques and particular reference to the needs AIDS? medical applications of ART. -
MF2574 Tips for a Successful Estrus Synchronization and Artificial
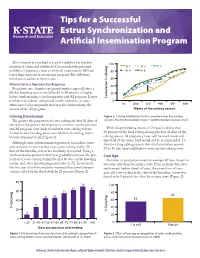
Tips for a Successful Estrus Synchronization and Artificial Insemination Program Do you know if your herd is a good candidate for synchro- 100 Yr 1 Yr 2 Yr 3 nization of estrus and ovulation? Can you identify potential problems if pregnancy rates to artificial insemination (AI) are 80 Yr 4 Yr 5 lower than expected in an existing program? The following information addresses these issues. 60 Natural Service Reproductive Response 40 22% Pregnancy rates (number pregnant/number exposed) after a 60-day breeding season should be 85 to 90 percent or higher 20 before implementing a synchronization and AI program. Lower % claving Cumulative 0 fertility may indicate suboptimal health, nutrition, or some 1st 2nd 3rd 6th5th4th other aspect of management that may be compromising the Weeks of the calving season success of the AI program. Calving Distribution Figure 2. Calving distribution for the same herd over five calving seasons. No synchronization in year 1; synchronization in years 2 to 5. The greater the proportion of cows calving the first 21 days of the season, the greater the response to an estrus synchronization and AI program. One study found that cows calving at least With longer breeding seasons (>70 days) and less than 71 days before breeding were more likely to be cycling and to 60 percent of the herd calving during the first 42 days of the become pregnant to AI (Figure 1). calving season, AI pregnancy rates, will be much lower and timed AI of the entire herd would not be recommended. To Although some synchronization protocols can induce estrus shorten a long calving season, two synchronization periods and ovulation in non-cycling cows, cows calving within 30 20 to 30 days apart will help to move up late-calving cows. -

Interferon-Tau Exerts Direct Prosurvival and Antiapoptotic
www.nature.com/scientificreports OPEN Interferon-Tau Exerts Direct Prosurvival and Antiapoptotic Actions in Luteinized Bovine Received: 13 May 2019 Accepted: 23 September 2019 Granulosa Cells Published: xx xx xxxx Raghavendra Basavaraja1, Senasige Thilina Madusanka1, Jessica N. Drum 2, Ketan Shrestha1, Svetlana Farberov1, Milo C. Wiltbank3, Roberto Sartori2 & Rina Meidan 1 Interferon-tau (IFNT), serves as a signal to maintain the corpus luteum (CL) during early pregnancy in domestic ruminants. We investigated here whether IFNT directly afects the function of luteinized bovine granulosa cells (LGCs), a model for large-luteal cells. Recombinant ovine IFNT (roIFNT) induced the IFN-stimulated genes (ISGs; MX2, ISG15, and OAS1Y). IFNT induced a rapid and transient (15– 45 min) phosphorylation of STAT1, while total STAT1 protein was higher only after 24 h. IFNT treatment elevated viable LGCs numbers and decreased dead/apoptotic cell counts. Consistent with these efects on cell viability, IFNT upregulated cell survival proteins (MCL1, BCL-xL, and XIAP) and reduced the levels of gamma-H2AX, cleaved caspase-3, and thrombospondin-2 (THBS2) implicated in apoptosis. Notably, IFNT reversed the actions of THBS1 on cell viability, XIAP, and cleaved caspase-3. Furthermore, roIFNT stimulated proangiogenic genes, including FGF2, PDGFB, and PDGFAR. Corroborating the in vitro observations, CL collected from day 18 pregnant cows comprised higher ISGs together with elevated FGF2, PDGFB, and XIAP, compared with CL derived from day 18 cyclic cows. This study reveals that IFNT activates diverse pathways in LGCs, promoting survival and blood vessel stabilization while suppressing cell death signals. These mechanisms might contribute to CL maintenance during early pregnancy.