Transcriptomic and Proteomic Analysis of Human Hepatic Stellate Cells Treated with Natural Taurine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genomic Structure and Expression of MJD, the Machado-Joseph Disease Gene
J Hum Genet (2001) 46:413–422 © Jpn Soc Hum Genet and Springer-Verlag 2001 ORIGINAL ARTICLE Yaeko Ichikawa · Jun Goto · Masahira Hattori Atsushi Toyoda · Kazuo Ishii · Seon-Yong Jeong Hideji Hashida · Naoki Masuda · Katsuhisa Ogata Fumio Kasai · Momoki Hirai · Patrícia Maciel Guy A. Rouleau · Yoshiyuki Sakaki · Ichiro Kanazawa The genomic structure and expression of MJD, the Machado-Joseph disease gene Received: March 7, 2001 / Accepted: April 17, 2001 Abstract Machado-Joseph disease (MJD) is an autosomal relative to the MJD gene in B445M7 indicate that there are dominant neurodegenerative disorder that is clinically char- three alternative splicing sites and eight polyadenylation acterized by cerebellar ataxia and various associated symp- signals in MJD that are used to generate the differently toms. The disease is caused by an unstable expansion of the sized transcripts. CAG repeat in the MJD gene. This gene is mapped to chromosome 14q32.1. To determine its genomic structure, Key words Machado-Joseph disease (MJD) · 14q32.1 · we constructed a contig composed of six cosmid clones and CAG repeat · Genome structure · Alternative splicing · eight bacterial artificial chromosome (BAC) clones. It spans mRNA expression approximately 300kb and includes MJD. We also deter- mined the complete sequence (175,330bp) of B445M7, a human BAC clone that contains MJD. The MJD gene was found to span 48,240bp and to contain 11 exons. Northern Introduction blot analysis showed that MJD mRNA is ubiquitously expressed in human tissues, and in at least four different Machado-Joseph disease (MJD) is an autosomal dominant sizes; namely, 1.4, 1.8, 4.5, and 7.5kb. -
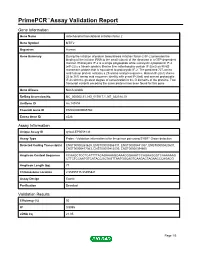
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name mitochondrial translational initiation factor 2 Gene Symbol MTIF2 Organism Human Gene Summary During the initiation of protein biosynthesis initiation factor-2 (IF-2) promotes the binding of the initiator tRNA to the small subunit of the ribosome in a GTP-dependent manner. Prokaryotic IF-2 is a single polypeptide while eukaryotic cytoplasmic IF-2 (eIF-2) is a trimeric protein. Bovine liver mitochondria contain IF-2(mt) an 85-kD monomeric protein that is equivalent to prokaryotic IF-2. The predicted 727-amino acid human protein contains a 29-amino acid presequence. Human IF-2(mt) shares 32 to 38% amino acid sequence identity with yeast IF-2(mt) and several prokaryotic IF-2s with the greatest degree of conservation in the G domains of the proteins. Two transcript variants encoding the same protein have been found for this gene. Gene Aliases Not Available RefSeq Accession No. NC_000002.11, NG_017017.1, NT_022184.15 UniGene ID Hs.149894 Ensembl Gene ID ENSG00000085760 Entrez Gene ID 4528 Assay Information Unique Assay ID qHsaCEP0058136 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000263629, ENST00000366137, ENST00000441307, ENST00000420637, ENST00000417363, ENST00000412530, ENST00000394600 Amplicon Context Sequence CCAAGCTCCTCATTTTACAGAAAAGGAAACGGAAATCCAGAAGGGTCAAAAAAG CTTCTCCAATGTCATACCGCTAGTTAATGGCAGTCAAGACTAGAACCCAGACG Amplicon Length (bp) 77 Chromosome Location 2:55495715-55495821 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 95 R2 0.9995 cDNA Cq 21.05 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79 gDNA Cq 24.56 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

Role of Phytochemicals in Colon Cancer Prevention: a Nutrigenomics Approach
Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Marjan J van Erk Promotor: Prof. Dr. P.J. van Bladeren Hoogleraar in de Toxicokinetiek en Biotransformatie Wageningen Universiteit Co-promotoren: Dr. Ir. J.M.M.J.G. Aarts Universitair Docent, Sectie Toxicologie Wageningen Universiteit Dr. Ir. B. van Ommen Senior Research Fellow Nutritional Systems Biology TNO Voeding, Zeist Promotiecommissie: Prof. Dr. P. Dolara University of Florence, Italy Prof. Dr. J.A.M. Leunissen Wageningen Universiteit Prof. Dr. J.C. Mathers University of Newcastle, United Kingdom Prof. Dr. M. Müller Wageningen Universiteit Dit onderzoek is uitgevoerd binnen de onderzoekschool VLAG Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Marjan Jolanda van Erk Proefschrift ter verkrijging van graad van doctor op gezag van de rector magnificus van Wageningen Universiteit, Prof.Dr.Ir. L. Speelman, in het openbaar te verdedigen op vrijdag 1 oktober 2004 des namiddags te vier uur in de Aula Title Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Author Marjan Jolanda van Erk Thesis Wageningen University, Wageningen, the Netherlands (2004) with abstract, with references, with summary in Dutch ISBN 90-8504-085-X ABSTRACT Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Specific food compounds, especially from fruits and vegetables, may protect against development of colon cancer. In this thesis effects and mechanisms of various phytochemicals in relation to colon cancer prevention were studied through application of large-scale gene expression profiling. Expression measurement of thousands of genes can yield a more complete and in-depth insight into the mode of action of the compounds. -

Low Abundance of the Matrix Arm of Complex I in Mitochondria Predicts Longevity in Mice
ARTICLE Received 24 Jan 2014 | Accepted 9 Apr 2014 | Published 12 May 2014 DOI: 10.1038/ncomms4837 OPEN Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice Satomi Miwa1, Howsun Jow2, Karen Baty3, Amy Johnson1, Rafal Czapiewski1, Gabriele Saretzki1, Achim Treumann3 & Thomas von Zglinicki1 Mitochondrial function is an important determinant of the ageing process; however, the mitochondrial properties that enable longevity are not well understood. Here we show that optimal assembly of mitochondrial complex I predicts longevity in mice. Using an unbiased high-coverage high-confidence approach, we demonstrate that electron transport chain proteins, especially the matrix arm subunits of complex I, are decreased in young long-living mice, which is associated with improved complex I assembly, higher complex I-linked state 3 oxygen consumption rates and decreased superoxide production, whereas the opposite is seen in old mice. Disruption of complex I assembly reduces oxidative metabolism with concomitant increase in mitochondrial superoxide production. This is rescued by knockdown of the mitochondrial chaperone, prohibitin. Disrupted complex I assembly causes premature senescence in primary cells. We propose that lower abundance of free catalytic complex I components supports complex I assembly, efficacy of substrate utilization and minimal ROS production, enabling enhanced longevity. 1 Institute for Ageing and Health, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 2 Centre for Integrated Systems Biology of Ageing and Nutrition, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 3 Newcastle University Protein and Proteome Analysis, Devonshire Building, Devonshire Terrace, Newcastle upon Tyne NE1 7RU, UK. Correspondence and requests for materials should be addressed to T.v.Z. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Genetic and Pharmacological Approaches to Preventing Neurodegeneration
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Genetic and Pharmacological Approaches to Preventing Neurodegeneration Marco Boccitto University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Neuroscience and Neurobiology Commons Recommended Citation Boccitto, Marco, "Genetic and Pharmacological Approaches to Preventing Neurodegeneration" (2012). Publicly Accessible Penn Dissertations. 494. https://repository.upenn.edu/edissertations/494 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/494 For more information, please contact [email protected]. Genetic and Pharmacological Approaches to Preventing Neurodegeneration Abstract The Insulin/Insulin-like Growth Factor 1 Signaling (IIS) pathway was first identified as a major modifier of aging in C.elegans. It has since become clear that the ability of this pathway to modify aging is phylogenetically conserved. Aging is a major risk factor for a variety of neurodegenerative diseases including the motor neuron disease, Amyotrophic Lateral Sclerosis (ALS). This raises the possibility that the IIS pathway might have therapeutic potential to modify the disease progression of ALS. In a C. elegans model of ALS we found that decreased IIS had a beneficial effect on ALS pathology in this model. This beneficial effect was dependent on activation of the transcription factor daf-16. To further validate IIS as a potential therapeutic target for treatment of ALS, manipulations of IIS in mammalian cells were investigated for neuroprotective activity. Genetic manipulations that increase the activity of the mammalian ortholog of daf-16, FOXO3, were found to be neuroprotective in a series of in vitro models of ALS toxicity. -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

Apoptotic Genes As Potential Markers of Metastatic Phenotype in Human Osteosarcoma Cell Lines
17-31 10/12/07 14:53 Page 17 INTERNATIONAL JOURNAL OF ONCOLOGY 32: 17-31, 2008 17 Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines CINZIA ZUCCHINI1, ANNA ROCCHI2, MARIA CRISTINA MANARA2, PAOLA DE SANCTIS1, CRISTINA CAPANNI3, MICHELE BIANCHINI1, PAOLO CARINCI1, KATIA SCOTLANDI2 and LUISA VALVASSORI1 1Dipartimento di Istologia, Embriologia e Biologia Applicata, Università di Bologna, Via Belmeloro 8, 40126 Bologna; 2Laboratorio di Ricerca Oncologica, Istituti Ortopedici Rizzoli; 3IGM-CNR, Unit of Bologna, c/o Istituti Ortopedici Rizzoli, Via di Barbiano 1/10, 40136 Bologna, Italy Received May 29, 2007; Accepted July 19, 2007 Abstract. Metastasis is the most frequent cause of death among malignant primitive bone tumor, usually developing in children patients with osteosarcoma. We have previously demonstrated and adolescents, with a high tendency to metastasize (2). in independent experiments that the forced expression of Metastases in osteosarcoma patients spread through peripheral L/B/K ALP and CD99 in U-2 OS osteosarcoma cell lines blood very early and colonize primarily the lung, and later markedly reduces the metastatic ability of these cancer cells. other skeleton districts (3). Since disseminated hidden micro- This behavior makes these cell lines a useful model to assess metastases are present in 80-90% of OS patients at the time the intersection of multiple and independent gene expression of diagnosis, the identification of markers of invasiveness signatures concerning the biological problem of dissemination. and metastasis forms a target of paramount importance in With the aim to characterize a common transcriptional profile planning the treatment of osteosarcoma lesions and enhancing reflecting the essential features of metastatic behavior, we the prognosis. -

By Submitted in Partial Satisfaction of the Requirements for Degree of in In
BCL6 maintains thermogenic capacity of brown adipose tissue during dormancy by Vassily Kutyavin DISSERTATION Submitted in partial satisfaction of the requirements for degree of DOCTOR OF PHILOSOPHY in Biomedical Sciences in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, SAN FRANCISCO Approved: ______________________________________________________________________________Eric Verdin Chair ______________________________________________________________________________Ajay Chawla ______________________________________________________________________________Ethan Weiss ______________________________________________________________________________ ______________________________________________________________________________ Committee Members Copyright 2019 by Vassily Kutyavin ii Dedicated to everyone who has supported me during my scientific education iii Acknowledgements I'm very grateful to my thesis adviser, Ajay Chawla, for his mentorship and support during my dissertation work over the past five years. Throughout my time in his lab, I was always able to rely on his guidance, and his enthusiasm for science was a great source of motivation. Even when he was traveling, he could easily be reached for advice by phone or e- mail. I am particularly grateful for his help with writing the manuscript, which was probably the most challenging aspect of graduate school for me. I am also very grateful to him for helping me find a postdoctoral fellowship position. Ajay's inquisitive and fearless approach to science have been a great inspiration to me. In contrast to the majority of scientists who focus narrowly on a specific topic, Ajay pursued fundamental questions across a broad range of topics and was able to make tremendous contributions. My experience in his lab instilled in me a deep appreciation for thinking about the entire organism from an evolutionary perspective and focusing on the key questions that escape the attention of the larger scientific community. As I move forward in my scientific career, there is no doubt that I will rely on him as a role model. -

B Inhibition in a Mouse Model of Chronic Colitis1
The Journal of Immunology Differential Expression of Inflammatory and Fibrogenic Genes and Their Regulation by NF-B Inhibition in a Mouse Model of Chronic Colitis1 Feng Wu and Shukti Chakravarti2 Fibrosis is a major complication of chronic inflammation, as seen in Crohn’s disease and ulcerative colitis, two forms of inflam- matory bowel diseases. To elucidate inflammatory signals that regulate fibrosis, we investigated gene expression changes under- lying chronic inflammation and fibrosis in trinitrobenzene sulfonic acid-induced murine colitis. Six weekly 2,4,6-trinitrobenzene sulfonic acid enemas were given to establish colitis and temporal gene expression patterns were obtained at 6-, 8-, 10-, and 12-wk time points. The 6-wk point, TNBS-w6, was the active, chronic inflammatory stage of the model marked by macrophage, neu- trophil, and CD3؉ and CD4؉ T cell infiltrates in the colon, consistent with the idea that this model is T cell immune response driven. Proinflammatory genes Cxcl1, Ccl2, Il1b, Lcn2, Pla2g2a, Saa3, S100a9, Nos2, Reg2, and Reg3g, and profibrogenic extra- cellular matrix genes Col1a1, Col1a2, Col3a1, and Lum (lumican), encoding a collagen-associated proteoglycan, were up-regulated at the active/chronic inflammatory stages. Rectal administration of the NF-B p65 antisense oligonucleotide reduced but did not abrogate inflammation and fibrosis completely. The antisense oligonucleotide treatment reduced total NF-B by 60% and down- regulated most proinflammatory genes. However, Ccl2, a proinflammatory chemokine known to promote fibrosis, was not down- regulated. Among extracellular matrix gene expressions Lum was suppressed while Col1a1 and Col3a1 were not. Thus, effective treatment of fibrosis in inflammatory bowel disease may require early and complete blockade of NF-B with particular attention to specific proinflammatory and profibrogenic genes that remain active at low levels of NF-B. -

Stroke Genetics and Genomics
Faculdade de Medicina da Universidade de Lisboa Unidade Neurológica de Investigação Clínica PhD Thesis Stroke Genetics and Genomics Tiago Krug Coelho Host Institution: Instituto Gulbenkian de Ciência Supervisor at Instituto Gulbenkian de Ciência: Doctor Sofia Oliveira Supervisor at Faculdade de Medicina da Universidade de Lisboa: Professor José Ferro PhD in Biomedical Sciences Specialization in Neurosciences 2010 Stroke Genetics and Genomics A ciência tem, de facto, um único objectivo: a verdade. Não esgota perfeitamente a sua tarefa se não descobre a causa do todo. Chiara Lubich i Stroke Genetics and Genomics ii Stroke Genetics and Genomics A impressão desta dissertação foi aprovada pela Comissão Coordenadora do Conselho Científico da Faculdade de Medicina de Lisboa em reunião de 28 de Setembro de 2010. iii Stroke Genetics and Genomics iv Stroke Genetics and Genomics As opiniões expressas são da exclusiva responsabilidade do seu autor. v Stroke Genetics and Genomics vi Stroke Genetics and Genomics Abstract ABSTRACT This project presents a comprehensive approach to the identification of new genes that influence the risk for developing stroke. Stroke is the leading cause of death in Portugal and the third leading cause of death in the developed world. It is even more disabling than lethal, and the persistent neurological impairment and physical disability caused by stroke have a very high socioeconomic cost. Moreover, the number of affected individuals is expected to increase with the current aging of the population. Stroke is a “brain attack” cutting off vital blood and oxygen to the brain cells and it is a complex disease resulting from environmental and genetic factors.